Figure 4.
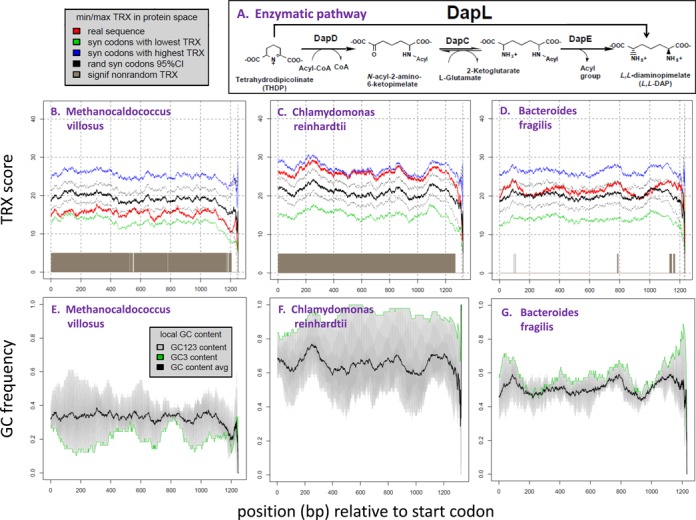
The intrinsic DNA flexibility and GC3 of the diaminopimelate aminotransferase (DapL) gene from three representative species. (A) Reaction catalyzed by DapL showing the ability of the enzyme to circumvent the DapD, DapC and DapE steps in the Escherichia coli acyl pathways. (DapD), N-acyl-2-amino-6-ketopimelate aminotransferase (DapC), N-acyl-L,L-2,6-ketopimelate deacylase (DapE), L,L-diaminopimelate aminotransferase (DapL) (46). Example plots of smoothed flexibility (TRX) scores (B–D) are shown for (B) an Archean, Methanonocaldococcus villosus,(C) an algae, Chlamydomonas reinhardtii and (D) a bacteria, Bacteroides fragilis. Plots B and C show DapL gene TRX scores (red) that are extreme examples of inflexible and flexible genes (respectively) for their given protein space (bounded in blue and green). The black lines indicate the average synonymous TRX score and 95% CI when synonymous codons are chosen randomly (local gene regions where observed TRX is outside this CI are flagged in brown on the X axis). A dapL gene's deviation from average synonymous flexibility is achieved largely by codon usage bias. Figures E–G show respective smoothed trends in overall GC content (black), GC content in each reading frame (gray) and GC3 (green).
