Figure 7.
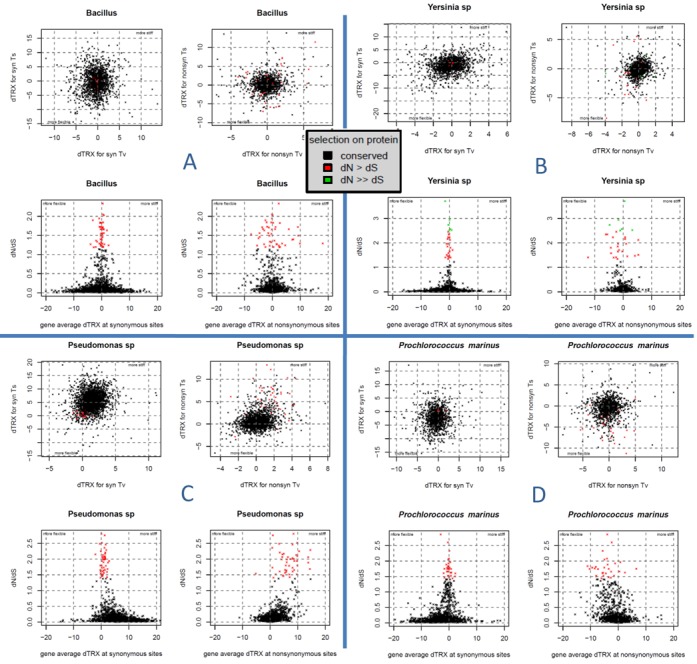
Fundamental constraints between protein evolution (dN/dS) and mutational impacts on intrinsic DNA flexibility (dTRX) and hence, genome architectural dynamics. Example plot sets are shown for two genomes with low codon bias (A) Bacillus and (B) Yersinia, and two genomes with extreme codon bias; (C) Pseudomonas = high GC and (D) Prochlorococcus = low GC. Within each plot set, genes functionally conserved at the protein level (i.e. low dN/dS) are shown in black, while genes adaptively altered at protein level are shown colored. Mutational impacts on flexibility (dTRX) are shown separately for synonymous sites (left side of plot set) and non-synonymous sites (right side of plot set). dTRX for transitions and transversions are separated in the upper plots of each set.
