Figure 3.
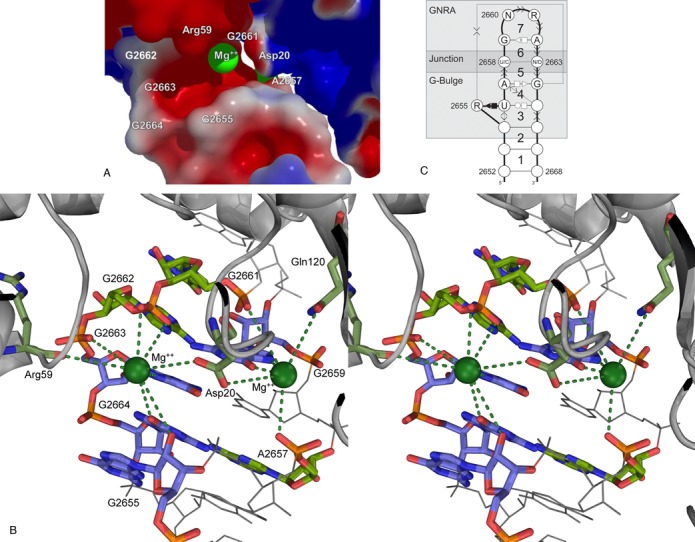
Structural determinants of viable bacterial SRLs. (A) Electrostatic surface of the sarcin–ricin/EF-G interaction. Surface potentials are presented in kT/e. The red color indicates negatively charged areas (EF-G from −1; SRL from −8) and blue positively charged areas (EF-G to 1; SRL to 0). The SRL/EF-G recognition put in close contact two negatively charged areas, made possible by the presence of two Mg2+ ions (green spheres). (B) Plausible Mg2+ coordination between the SRL and EF-G from crystal structures (PDB IDs 4KIX and 4KIY). The green dashed lines indicate possible direct coordination. One of the Mg2+ ions (green sphere on the right) is located near atoms Gln120:OE1 and Asp20:OD2 of the EF-G, and A2657:O2P, G2659:O6, G2659:N7 and G2661:O1P of the SRL. The second Mg2+ ion (green sphere on the left) is near atoms Arg59:O and Asp20:OD1 of the EF-G, and G2655:O2′, G2662:O1P, G2662:O2P, G2663:N7 and G2664:O6 of the SRL. The EF-G residues and SRL nucleotides involved in Mg2+ coordination are shown with colored sticks; others in gray. The bases G2655, G2659, G2663 and G2664 stack and are shown with blue carbons. (C) The SRL structural determinants represented as sequence and base interaction constraints. The SRL is composed of the GNRA and G-bulge motifs linked by the Y2658:N2663, but C2658:C2663, junction base pair.
