Figure 4.
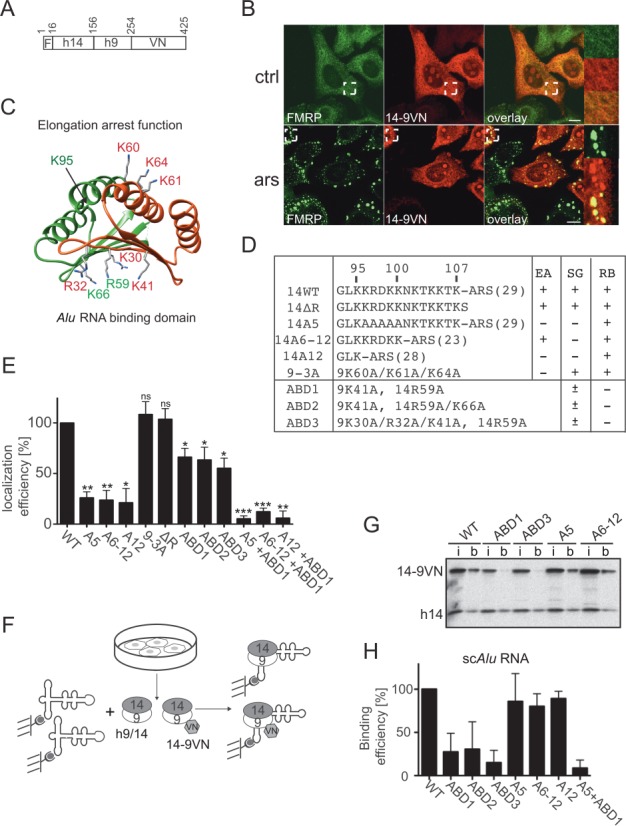
Functional determinants for SG localization in SRP9/14. (A) Schematic representation of the 14-9VN fusion protein. F: flag epitope: VN: 173 N-terminal amino acid residues of the Venus protein. Numbering refers to amino acid residues in 14-9VN. (B) Double immunofluorescence staining of HeLa cells with anti-GFP and anti-FMRP antibodies. ars: 500 μM sodium arsenite for 30 min; ctrl: untreated cells. Images were captured using a 63x lens on the LSM-710 laser scanning microscope. Areas denoted by rectangles are shown at higher magnification. Scale bars: 10 μm. (C) Structure of the protein dimer. h9: red, h14: green. Mutated amino acids are highlighted. The structure of the protein sequence following K95 in h14 could not be solved in the SRP-Alu-h9/14 complex (32). (D) Description of the mutations in the reporter protein 14-9VN. Amino acids are numbered according to the human sequences. ARS: alanine-rich sequence; brackets: number of amino acid residues; EA: elongation arrest activity; SG: SG localization; RB: Alu RNA binding activity. ABD1–3: proteins with mutations in the Alu RNA Binding Domain. (E) Efficiency of SG localization of the mutated proteins. Wild-type (WT) and mutated reporter proteins were expressed in HeLa Kyoto cells. Double immunofluorescence staining of cells with antibodies against GFP and FMRP revealed the fusion proteins and SGs, respectively. The presence of the reporter protein in SGs was counted in 100 transfected and SG-positive cells. In 74 ± 10% (n = 8) of these cells, 14-9VN was present in SGs. Localization efficiencies of the mutated proteins were normalized to 14-9VN, which was arbitrarily set to 100%. Error bars are shown as SD, n = 3. (F) Schematic representation of the RNA binding assay. Synthetic biotinylated scAlu RNA immobilized on magnetic streptavidin beads was incubated with postnuclear supernatants of HeLa Kyoto cells expressing the fusion proteins. (G) Equivalent aliquots of the input (i) and scAlu RNA-bound (b) protein fractions were analyzed by Western blot. Additional Western blots of RNA binding experiments are shown in Supplementary Figure S4. (H) Quantification of the RNA binding efficiencies of all fusion proteins normalized to the WT, which was set to 100%. Error bars are shown as SD, n ≥ 2. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
