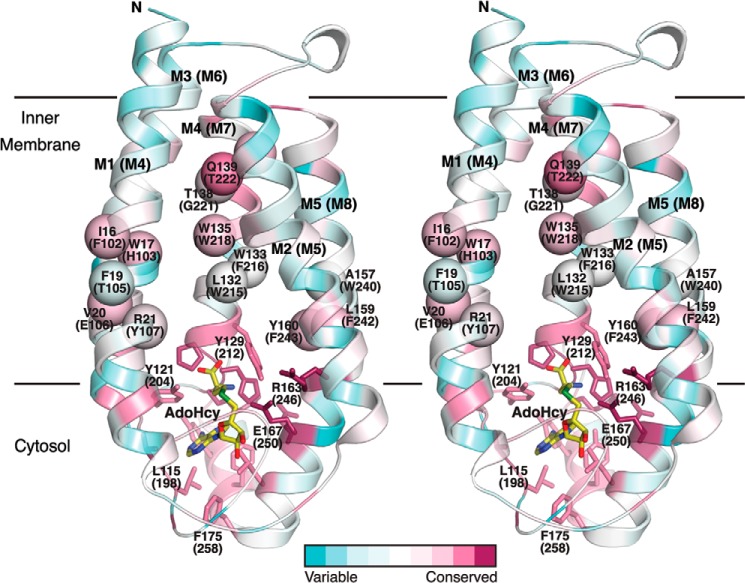
FIGURE 11.
Residues proposed to be involved in substrate binding in ICMT mapped on the structure of Ma MTase. The schematic representation of the Ma MTase structure (PDB code 4A2N) is colored according to amino acid sequence conservation with the ICMT family members (pink, high sequence conservation; green, low sequence conservation) in stereoview. The sequence alignment was made in ClustalW with manual adjustments according to Fig. 2, the conservation was analyzed and mapped on the structure using the ConSurf Server (44), and the figure was constructed with PyMOL. Amino acid numbering corresponds to Ma MTase, with Ag ICMT numbering in parentheses. Residues that contribute to AdoMet binding are drawn as sticks. The approximate locations of Ag ICMT residues in the transmembrane helices M4, M7, and M8 that our study suggests contribute to the isoprenylcysteine binding site (on the basis that mutants exhibit either substrate inhibition or are inactive) are indicated as spheres. AdoHcy is drawn as sticks and colored yellow.