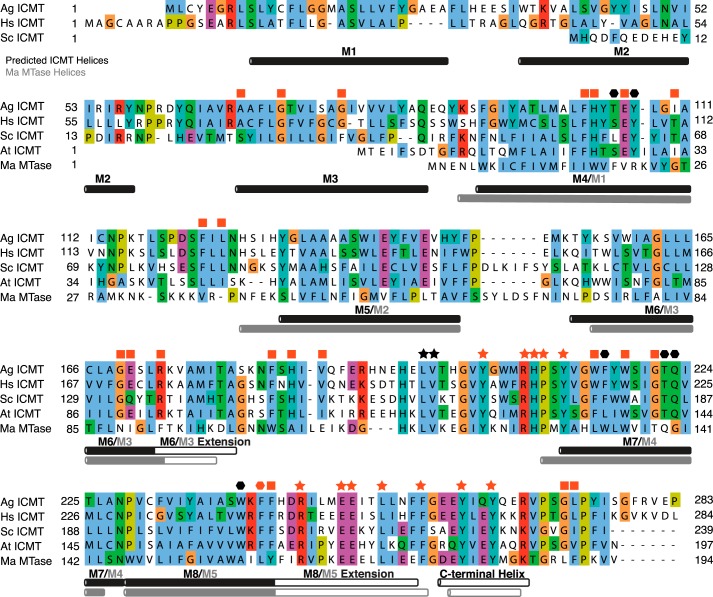
FIGURE 2.
Sequence alignment of ICMT orthologs and Ma MTase. Sequence alignment of Ag ICMT (283 amino acids (aa)), Hs ICMT (284 aa), Sc ICMT (239 aa), At ICMT (197 aa), and a prokaryotic integral membrane methyltransferase of known structure, Ma MTase (194 aa). Helices are indicated by horizontal rectangles below the aligned sequences. Helical regions of ICMT (black rectangles) were predicted using Jpred (43) and are also based on hydropathy plots and experimental mapping of Hs ICMT (19), where solid black rectangles represent regions predicted to reside in the membrane and open rectangles indicate helical regions in the cytosol. The Ma MTase helices (gray rectangles) and transmembrane helices (solid gray) were determined from the crystal structure (20). ICMT residues identified in this study as important for substrate binding are indicated: stars mark residues involved in AdoMet binding, and hexagons mark residues for which mutants have altered isoprenylcysteine substrate binding properties. All ICMT residues whose mutants were inactive are colored red; a red square indicates a residue that severely impaired activity when mutated but whose particular role has not been assigned. The UniProt accession numbers for the sequences in the alignment are: O60725, Q7PXA7 (version 2), P32584, Q93W54, and Q8TMG0. Alignment was made with ClustalW with manual adjustments. Conservation of the residues at the N terminus of Ag ICMT and Hs ICMT (M1 and M2) is poor, and as such, the alignment is less certain in this region.