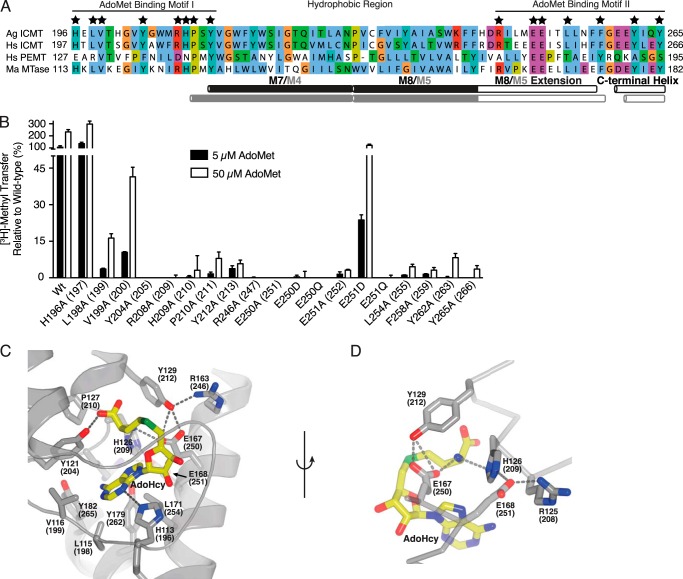
FIGURE 7.
Mutation of the AdoMet binding motif. A, sequence alignment of the conserved AdoMet binding region in Ag ICMT, Hs ICMT, Hs PEMT, and Ma MTase. Residues that interact with AdoHcy in the Ma MTase structure are indicated by a star. The UniProt accession number for Hs PEMT is CAG33380. Alignment was made in ClustalW with manual adjustments. B, mutational analysis of the residues conserved between Ma MTase and ICMT that coordinate AdoHcy in the Ma MTase structure. All residues were mutated to alanine. For Glu-250 and Glu-251, conservative substitutions to aspartic acid and glutamine were also evaluated. The activity assay was conducted using 4 μm BFC and 5 or 50 μm AdoMet. The data shown are the mean of duplicate measurements with error bars representing ± 1 S.D. Ag ICMT numbering is shown with Hs ICMT numbering in parentheses. C, details of the AdoMet binding site from the crystal structure of Ma MTase (PDB code 4A2N). AdoHcy and residues involved in binding AdoHcy (Ma MTase numbering with Ag ICMT numbering in parentheses) are drawn as sticks, and hydrogen bonds are indicated as dashed lines. D, details of the interaction of Glu-167 and Glu-168 (Glu-250 and Glu-251 in Ag ICMT) with AdoHcy. C and D were generated in PyMOL.