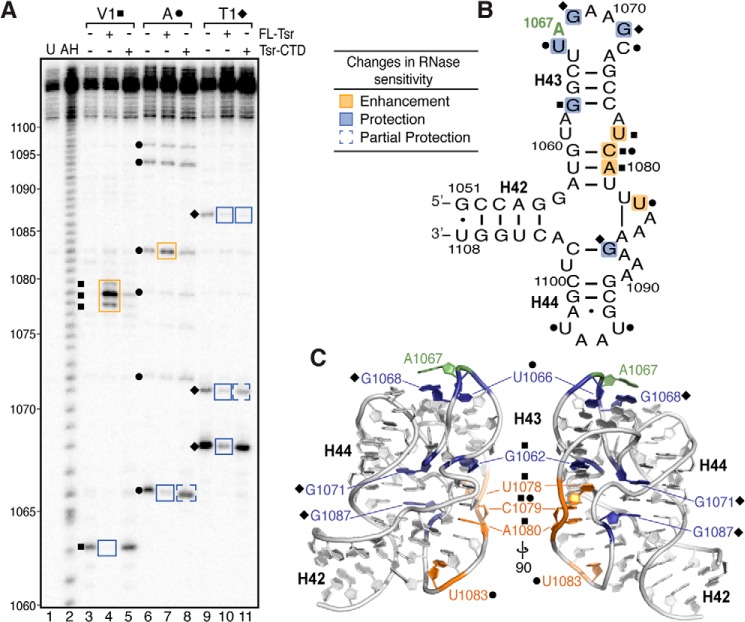
FIGURE 5.
RNase probing identifies RNA structural changes and cleavage protections induced upon binding full-length Tsr or Tsr-CTD. A, representative RNase structure probing gel of 58-nt RNA only, and complexes of 58-nt RNA with full-length Tsr or Tsr-CTD. Untreated RNA (U) and partial alkaline hydrolysis nucleotide ladder (AH) are also shown for nucleotide identification (numbering noted on left). Sites of cleavage are noted for RNase V1 (squares), RNase A (circles), RNase T1 (diamonds). Changes in sensitivity to RNases (boxed on the gel) are mapped as protections (blue) and enhancements (orange) onto B, the 58-nt RNA secondary structure, and C, two views of x-ray crystal structure (PDB ID 1HC8) related by a 90° rotation around the vertical axis.