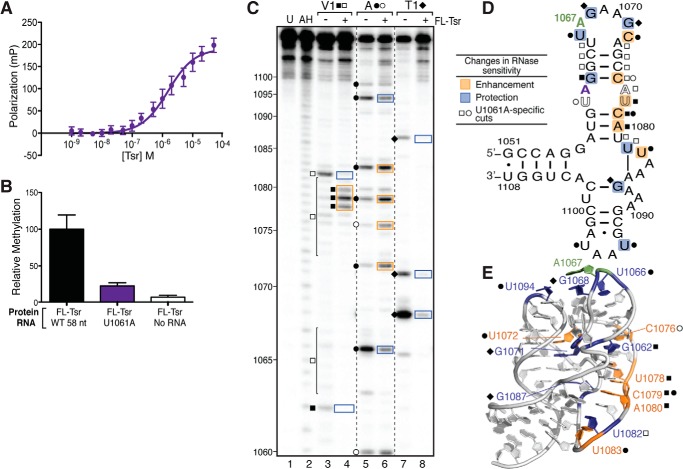
FIGURE 6.
Stabilizing the RNA tertiary structure decreases Tsr binding affinity and catalytic activity independent of RNA structural changes. A, FP binding analysis of U1061A RNA and full-length (FL) Tsr interaction. B, relative methylation activity of full-length Tsr on the wild-type (WT) and U1061A mutant 58-nt RNAs. C, representative RNase structure probing gel of U1061A RNA in the absence and presence of full-length Tsr. Untreated RNA (U) and partial alkaline hydrolysis nucleotide ladder (AH) are also shown for nucleotide identification (numbering noted on left). Sites of cleavage are noted for RNase V1 (squares), RNase A (circles), and RNase T1 (diamonds); cleavage sites unique to the U1061A mutant are shown as outlined symbols. Changes in sensitivity to RNase are mapped as protections (blue) and enhancements (orange) onto the 58-nt RNA. D, secondary structure, and E, x-ray crystal structure (PDB ID 1HC8).