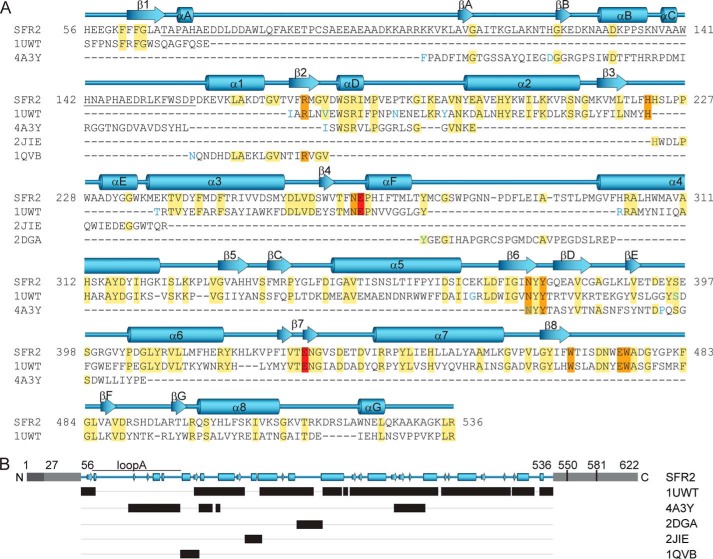
FIGURE 2.
Alignment of SFR2 and homolog sequences used to build the three-dimensional structural model of SFR2. Amino acid sequence (A) is shown with SFR2 sequence numbering. Other family 1 glycosyl hydrolases are indicated by Protein Data Bank identifiers and are from the following species: S. solfataricus, 1UWT; R. serpentina, 4A3Y; T. aggregans, 1QVB; P. polymyxa, 2JIE; and T. aestivum, 2DGA. For space reasons, aligned template (1UWT and 4A3Y) sequences with insertions resulting in gaps greater than one residue in the SFR2 sequence are not shown. These positions are indicated by blue coloring of the following residue. Identical residues are highlighted in yellow, active site residues in orange, and acid/base catalyst glutamates in red. Secondary structure of the model is displayed above the sequence with (β/α)8 barrel helices and strands numbered sequentially, as per glycosyl hydrolase conventions. Additional helices and strands are lettered sequentially. Loop A, which is referred to specifically in the text, is underlined. The schematic representation in B, shows in black bars the ranges of residues in different structures that were used to construct the SFR2 structural model. Gray regions indicate portions of SFR2 not included in the model. Light gray portions have no known function, and dark gray indicates an identified transmembrane domain. Residue numbers for key features mentioned in the text are noted.