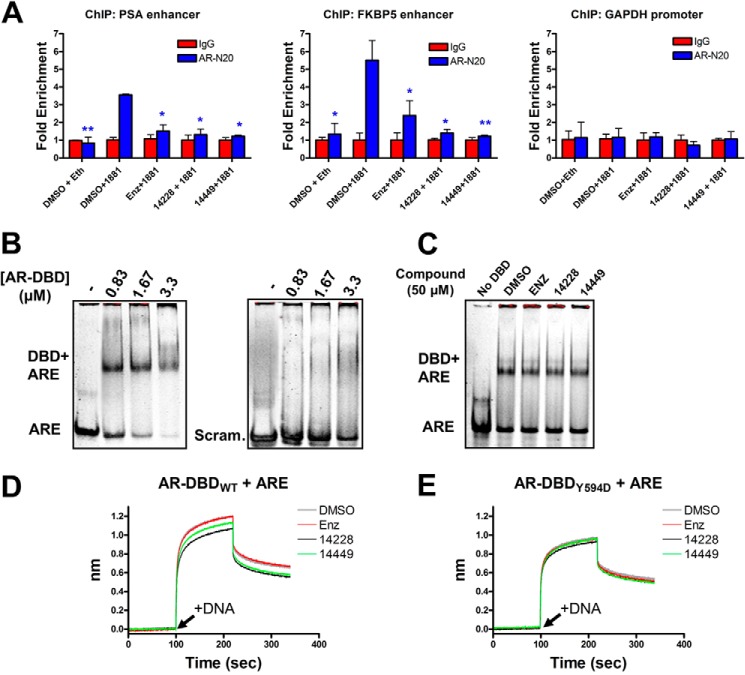
FIGURE 6.
Protein-DNA complexes are diminished in the presence of compounds. A, ChIP analysis of AR binding to the PSA or FKBP5 enhancers, or the GAPDH promoter, in LNCaP cells. Where indicated, nuclear translocation of the AR was stimulated with 1 nm R1881 (or ethanol only), and compounds were administered at 10 μm concentration. Sheared chromatin-protein complexes were precipitated with the AR-N20 antibody, reverse cross-linked, and analyzed by quantitative PCR. The results are normalized as fold enrichment over precipitation with a rabbit isotype control IgG antibody for each condition tested. Error bars represent the mean ± S.D. between three replicates. Fold enrichment from each tested condition (AR-N20 antibody) was statistically compared against DMSO + 1881 with a two-tailed t test (paired): *, p value < 0.05; **, p value < 0.01. B, purified AR-DBD + hinge domain at the indicated concentration was mixed with ∼2 μm ARE or scrambled 42-bp dsDNA and separated by native-PAGE. C, same as B but with 0.83 μm AR-DBD in the presence of 50 μm compound. D, biotinylated AR-DBD + hinge was loaded onto streptavidin sensors for biolayer interferometry analysis. t = 0 s. DBD was equilibrated with the indicated compound (50 μm); t = 100 s (arrow), dsDNA association was monitored with 3 μm ARE + 50 μm compound; t = 220 s, dissociation in buffer + compound. The y axis represents the nanometer shift in wavelength resulting from ligand (dsDNA) binding/dissociation. E, same as D but with the DBD bearing the Y594D mutation. enz, enzalutamide.