Figure 2.
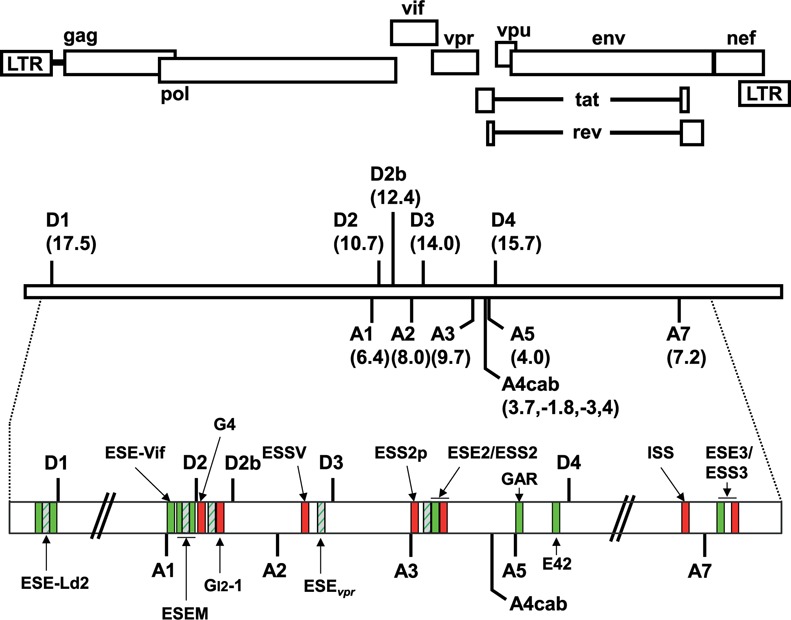
Schematic drawing of known and HEXplorer-predicted splicing regulatory elements (SREs) in the HIV-1 genome. Top: open reading frames are indicated by open boxes. The long terminal repeats (LTRs) are located at both ends of the provirus. Center: all HIV-1 proteins are encoded by a single primary RNA. Alternative splicing leads to more than 40 different mRNA transcripts enabling efficient translation of all open reading frames within the host cell. Intrinsic strength of the 5′ss (D1–D4) and 3′ss (A1–A7) is indicated in the parentheses (5′ss: HBond Score, http://www.uni-duesseldorf.de/rna; 3′ss: MaxEntScore, http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html). Bottom: positions of the SREs within the HIV-1 pre-mRNA: known splicing enhancers (green) and silencers (red) are indicated together with HEXplorer-predicted enhancers (dashed green). [ESE-Ld2 (25); ESE-Vif (27); ESEM (26); guanosine (G)-rich silencer G4 (27); GI2–1 (23); ESSV (19,21–22); ESEvpr (20); ESS2p (28); ESE2 (29,30); ESS2 (31–33); guanosine-adenosine-rich (GAR) ESE (24,34,46); E42 fragment (34); ISS (35); ESE3 (36); ESS3 (36–38) (adapted to (34,39)].
