Figure 4.
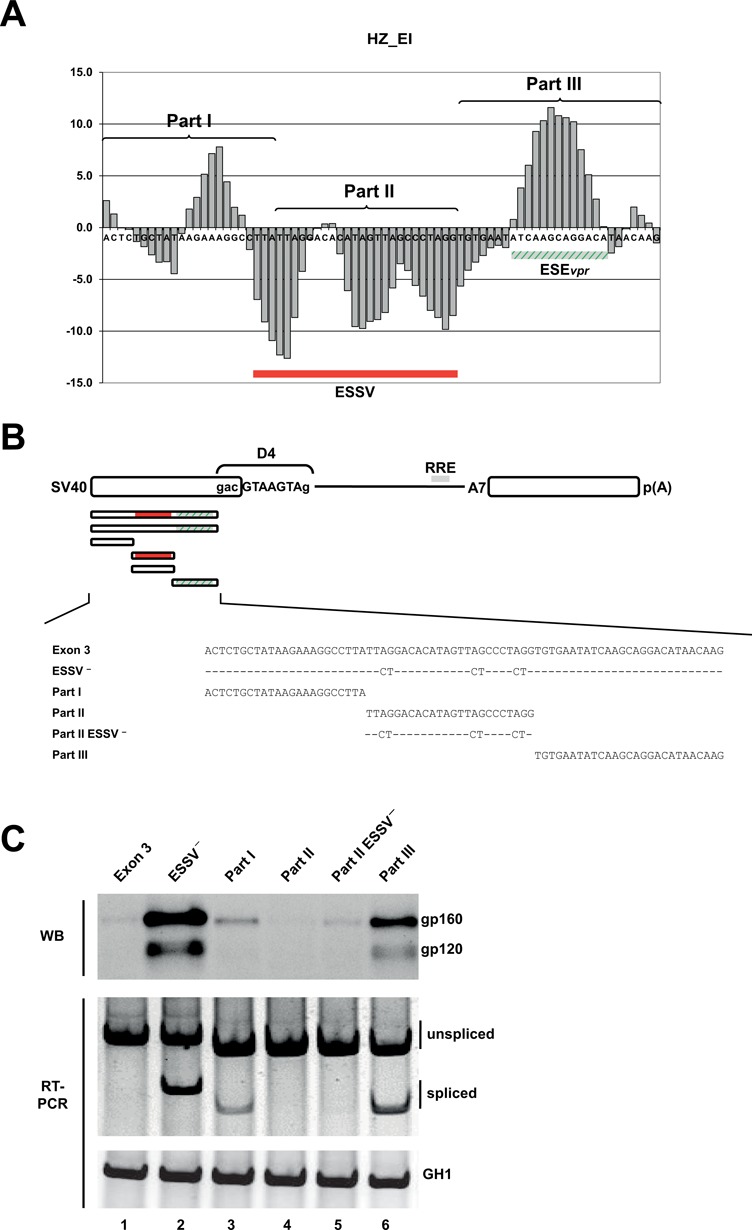
Splice enhancing and silencing properties of exon 3 fragments correlate with HEXplorer score. (A) HZEI-score plot of the HIV-1 exon 3 sequence partitioned into three consecutive fragments (parts I–III) of equal sequence length. HZEI-positive and -negative regions are indicated in green and red, respectively. (B) Schematic of the SV-env expression plasmid carrying the env open reading frame (ORF). HIV-1 exon 3-derived sequences that were inserted upstream of the splicing enhancer dependent 5′ss D4 are enlarged below. ESSV− is known to disrupt the silencer ESSV. Either single part I, part II, part II ESSV− or part III was inserted, but no combinations. (C) Western blot analysis of cell lysates from HeLa cells transiently transfected with 1 μg of each of the constructs together with 1-μg SVcrev and 1 μg of pGL3 (Promega) to control for equal transfection efficiencies. Forty-eight hours post transfection proteins were extracted and separated by 7% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Samples were normalized for equal protein and luciferase amounts as described previously (46). After transfer to a nitrocellulose membrane, samples were probed with a mouse monoclonal antibody specifically detecting splicing-dependent gp120 expression within the transfected cells (87–133/026; kindly provided by Dade Behring). (SV40: simian virus 40 early promoter; RRE: Rev responsive element.)
