Figure 5.
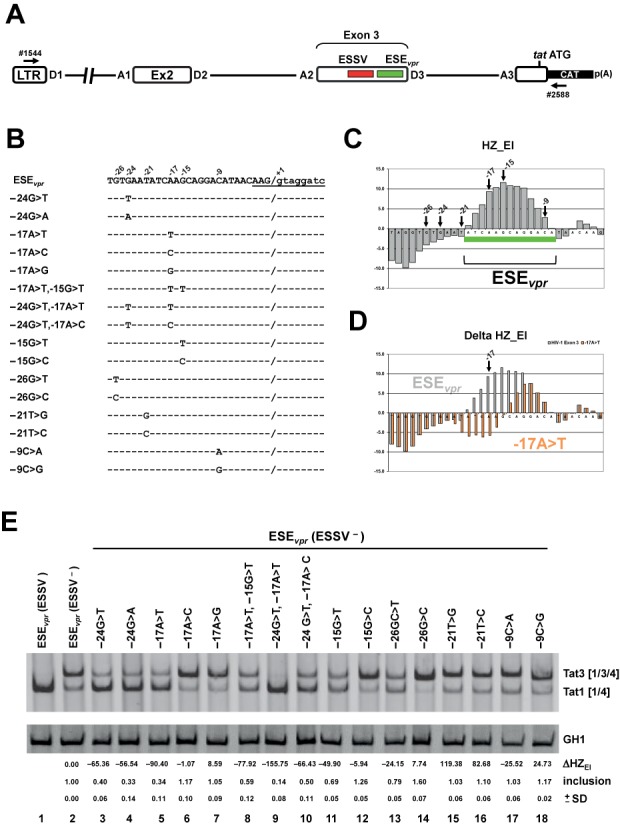
Detailed HEXplorer-guided mutational analysis of ESEvpr. (A) Schematic of the HIV-1-based LTR ex2 ex3 splicing reporter. Positions of the RT-PCR primers (#1544/#2588) are indicated by arrows. (B) ESEvpr reference (pNLA1) and mutant sequences. Nucleotide residues are denoted by their position relative to the GT-dinucleotide. Exon–intron border of exon 3 is denoted by ‘/’ and 5′ss D3 sequence is underlined. (C) HEXplorer score profile plotted along the 3’ end of exon 3 containing ESEvpr (green bar). Positions of mutated nucleotides are indicated by arrows. (D) HEXplorer score profiles for reference ESEvpr (gray) and exemplary point mutation −17A>T (red, indicated by arrow) plotted along the same sequence as in (C). (E) RT-PCR of RNA from HeLa cells transfected with mutants described in (B). HEXplorer score difference ΔHZEI between mutant and reference (ESEvpr (ESSV−) lane 2) as well as exon 3 inclusion (upper band Tat3 [1/3/4]) level is given below each lane. Mean inclusion level and standard deviation (±SD) were calculated from triplicates. GH1 (growth hormone) serves as control for transfection efficiency. 2.5 × 105 HeLa cells were transiently transfected with 1 μg of each of the constructs together with 0.2 μg of SVctat and 1 μg of pXGH5. Thirty hours after transfection RNA was isolated from the cells and subjected to RT-PCR analysis using primer pairs #1544/#2588 and #1224/#1225 (GH1). PCR products were separated by 8% non-denaturating polyacrylamide gel electrophoresis and stained with ethidium bromide.
