Figure 1.
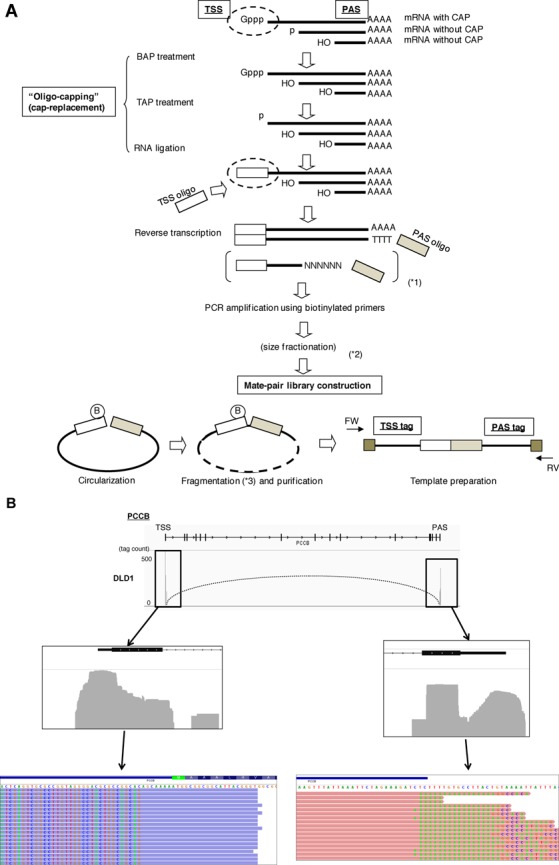
Construction and characterization of the mate pair full-length cDNA libraries. (A) Schematic representation of the mate pair full-length cDNA library construction. Briefly, the cap structure of RNA was replaced with a synthetic oligo-ribonucleotide by BAP-TAP-RNA ligase treatment. Full-length cDNAs with adaptor sites at both ends were amplified by PCR. Biotinylated-oligo was used for the 5′-end PCR primer. The PCR amplicons were circularized and fragmented. Subsequently, the fragments with biotin were recovered by avidin column. Non-circularized fragments were degraded by exonuclease treatment. For the purified ‘TSS-PAS’ mate pair fragments, sequence adaptors were ligated to the both ends, and templates for the next-generation sequencing were generated. Details of the protocols are described in the Materials and Methods section and Supplementary Methods as well as on our web site: http://dbtss.hgc.jp/cgi-bin/protocol_matepairLibrary.cgi. (B) Examples of TSCs and PACs identified in the TSS/PAS libraries. The case of the PCCB gene is shown. The peaks indicate the tag counts from the TSS/PAS mate pair libraries (upper). The numbers of tag counts are shown in the margin. The dotted curve indicates the TSC-PAC pair. The cell type from which the TSC/PAC was derived is shown in the left margin. Magnifications of the upper panels are shown in the middle panels for the indicated regions. Lower panels show the tag sequences. Bases derived from adaptor or poly(A) sequences, which were thus mismatched to the reference genome, are highlighted.
