Figure 1.
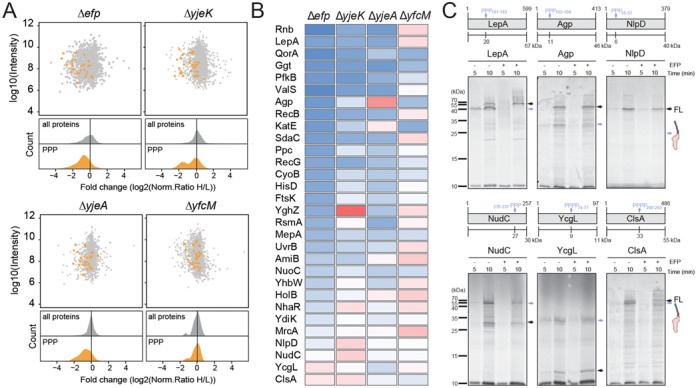
Proteomic analysis of BW25113 strains lacking efp, epmA, epmB and epmC. (A) Scatter and density plots of the inverted normalized H/L ratios (log2-transformed) relative to the summed-up protein intensity for a SILAC data from the Δefp, ΔyjeK, ΔyjeA and ΔyfcM for all proteins (grey) and PPP-containing proteins (orange). (B) Heat map representation of the 30 identified PPP-containing proteins that were up- (red) and down- (blue) regulated in the Δefp, ΔyjeK, ΔyjeA and ΔyfcM strains relative to wild-type strain. (C) In vitro translation time course of selected PPP-containing protein. Autoradiographs of SDS-PAGE and analysis of [35S]Met-labelled LepA, Agp, NlpD, NudC, YcgL and ClsA. All reactions were performed in the absence (−) and presence (+) of active EF-P. The position of FL protein and intermediate peptidyl-tRNA indicated with black and blue arrows, respectively. Schematic representations of each protein are included, with the site of the PPP motif indicated by a blue arrow.
