Figure 3.
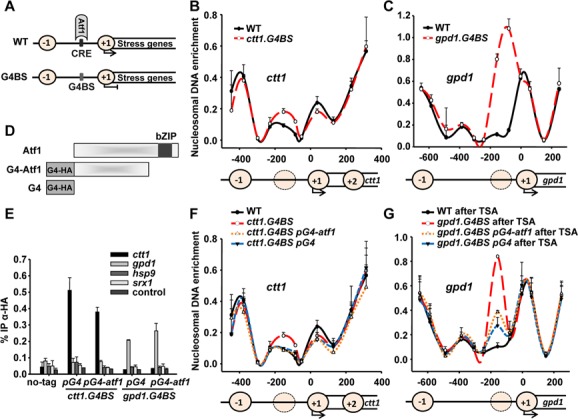
CRE site substitution by Gal4-binding site reduces NDR which is restored by artificially tethering Gal4-Atf1 or the Gal4 binding domain. (A) Schematic representation of CRE site substitution by a Gal4 binding site (G4BS). (B and C) Nucleosome-scanning analysis was performed as described in Figure 1B and C from cultures of strains 972 (WT) and EP184 (ctt1.G4BS; B) or EP255 (gpd1.G4BS; C). (D) Schematic representation of the fusion proteins Gal4-Atf11–395 (lacking the bZIP; G4-Atf1) and Gal4 DNA binding domain (G4). (E) Gal4 binding domain and Gal4-Atf11–395 physically bind to ctt1 or gpd1 promoters with a CRE-to-G4BS substitution. Cultures of strains 972 (no-tag), EP213 (ctt1.G4BS pG4), EP212.ΔbZIP (ctt1.G4BS pG4-atf1), EP286 (gpd1.G4BS pG4) and EP287.ΔbZIP (gpd1.G4BS pG4-atf1) were used to perform ChIP experiments using primers at ctt1, gpd1, hsp9 and srx1 promoters. An intergenic region was used as a negative control (control). Error bars (SEM) for all ChIP experiments were calculated from biological triplicates. (F) Mononucleosomes were isolated from cultures of strains 972 (WT), EP184 (ctt1.G4BS), EP213 (ctt1.G4BS pG4) and EP212.ΔbZIP (ctt1.G4BS pG4-atf1), and nucleosome scanning performed as described in Figure 1B. (G) Mononucleosomes were isolated from cultures of strains 972 (WT), EP255 (gpd1.G4BS), EP286 (gpd1.G4BS pGal4) and EP287.ΔbZIP (gpd1.G4BS pG4-atf1) treated with trichostatin A for eight generations, then washed and allowed to grow for eight generations without the drug; nucleosome scanning was then performed as described in Figure 1B. Error bars (SEM) were calculated from biological duplicates (B, C, F and G).
