Figure 4.
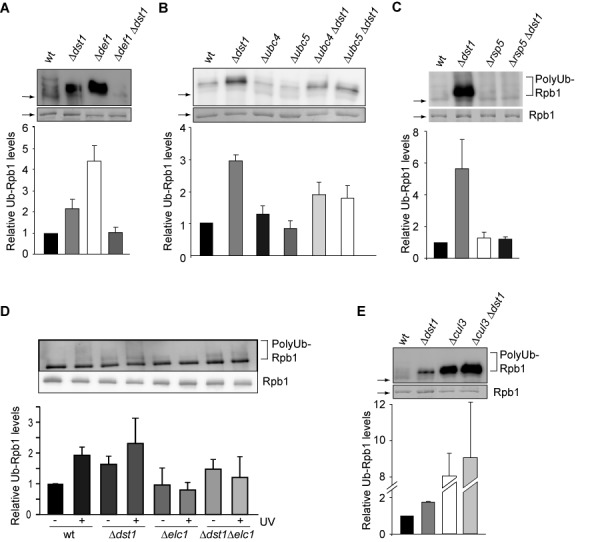
Mechanism of polyubiquitylation of Rpb1 of DNA damage-independently stalled RNAPII. (A–E) Polyubiquitylation of Rpb1 in Δdst1 and Δubc4, Δubc5, Δdef1, Δrsp5, Δelc1, Δcul3 and the respective double mutant strains was assessed after purification of approximately equal levels of RNAPII using Rpb3-TAP and limiting amounts of beads in the presence of DUB and proteasome inhibitors. Rpb1 levels were assessed by Coomassie staining and polyubiquitylation by western blotting using a ubiquitin specific antibody (upper panel). In (D) cells were also treated with UV (+UV). Note that Rpb1 ubiquitylation is unusually high in the Δrsp5 strain, which is probably due to the oleic acid and NP40 containing media needed for viability of the Δrsp5 strain. An explanation for the increased Rpb1 ubiquitylation in the Δcul3 and Δcul3Δdst1 strains could be that impairment of the E3 ligase Cul3 most likely leads to decreased ubiquitylation of Cul3-dependent substrates, which might lead to higher levels of free ubiquitin causing in turn unspecific ubiquitylation of Rpb1. However, there is no satisfactory explanation for the observed increase in Rpb1 ubiquitylation in the Δdef1 strain, which contradicts the current model. Columns and error bars represent the mean ± standard deviation from four independent experiments (bottom panels). P-values are listed in Supplementary Table S3.
