Figure 7.
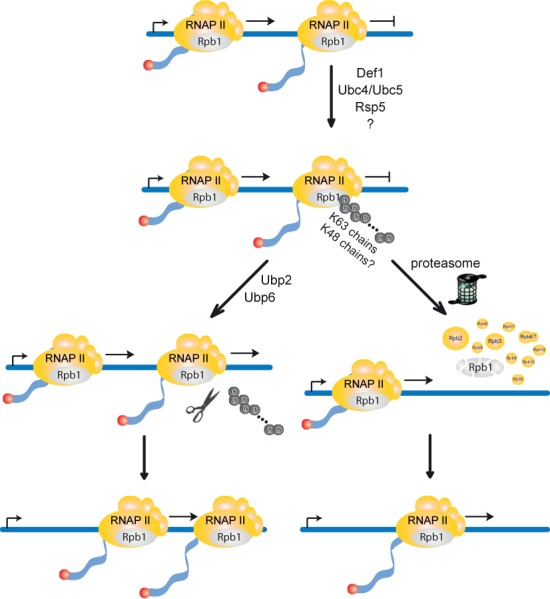
Model for the degradation of DNA damage-independently stalled RNA polymerase II (RNAPII). Persistently stalled RNAPII complexes are polyubiquitylated with K63-specific polyubiquitin chains by Def1, the E2s Ubc4 and Ubc5 and the E3 Rsp5. When transcription factors restart RNAPII, Rpb1 is deubiquitylated by Ubp2 or Ubp6 (left pathway) and RNAPII resumes transcription. Alternatively, as a failsafe mechanism when RNAPII remains persistently stalled, polyubiquitylated Rpb1 is recognized by the proteasome and degraded (right pathway). For Rpb1 degradation a remodeling of the K63-linked polyubiquitin chains and conjugation of K48-linked polyubiquitin chains by a yet to be identified E3 ligase might be necessary. This pathway for degradation of RNAPII complexes stalled persistently during transcription elongation frees the way for following RNAPII complexes. The two pathways for Rpb1 degradation, DNA damage-dependent and -independent, are compared in Supplementary Figure S1.
