Figure 5.
Regeneration Defects of Neural Cell Types following Tissue-Associated Transcription Factor RNAi
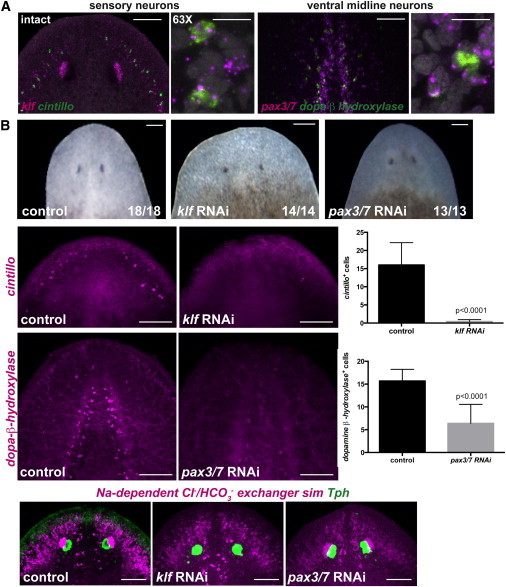
(A) Two regions of the brain are shown: the left panel depicts sensory neurons coexpressing the transcription factor klf and the marker cintillo, and the right panel shows ventral midline neurons coexpressing the transcription factor pax3/7 and the enzyme dopamine-β-hydroxylase (scale bars, 100 μm). Higher-magnification panels show coexpression of the genes analyzed in intact head regions (scale bars, 10 μm). DAPI labeled DNA (gray).
(B) Upper row: normal head regeneration (day seven) in control, klf, or pax3/7 RNAi animals. Animals were fed four times followed by head amputation. Second row: cintillo+ cells are not detected in klf(RNAi) animals; graph on the right shows total number of cintillo+ cells (mean ± SD) in regenerating heads (n > 14 animals per RNAi condition; Student’s t test, p < 0.0001). Third row: dopamine-β-hydroxylase-expressing cells are strongly reduced in regenerating pax3/7(RNAi) animals; graph on the right shows total number of dopamine-β-hydroxylase cells (mean ± SD) in regenerating heads (n > 13 animals per RNAi condition; Student-t test, p < 0.0001). Fourth row: dorsal view of control labelings using the RNA probes sim, the outer branches and photoreceptor neuron gene Na+-dependent Cl−/HCO3−, and the trypthophan hydroxylase (Tph) gene, show normal expression of these genes in the regenerating heads of the different RNAi animals. Images are maximal-intensity projections and representative of results seen in more than five animals (except otherwise labeled) per panel. Anterior, up. Scale bars, 100 μm.
