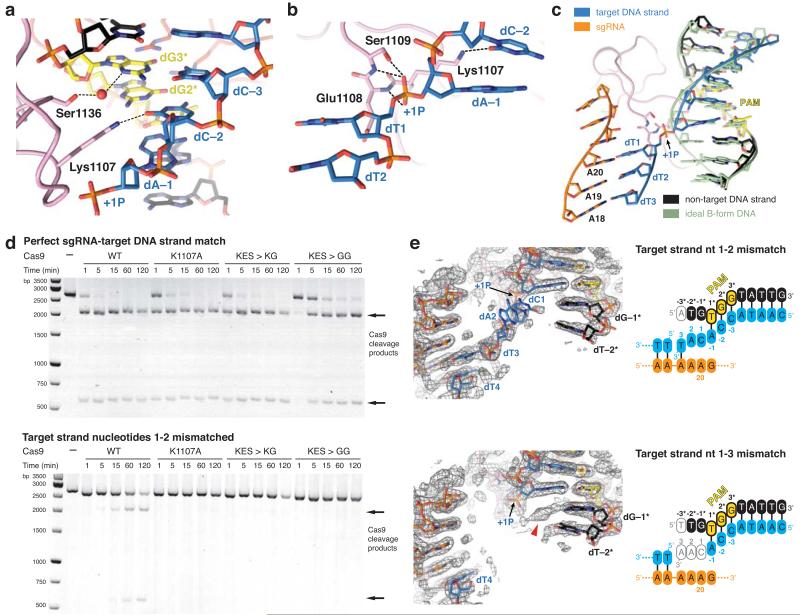
Figure 3. Interactions with the +1 phosphodiester group orient the target strand for guide RNA binding.
a, Detailed view of the minor groove of the PAM region. b, Hydrogen-bonding interactions (dashed lines) of the +1 phosphate (+1P) with the Lys1107-Ser1109 (phosphate lock) loop. c, Superposition of the unwound target DNA strand with an ideal B-form DNA duplex (green) d, Endonuclease activity assays using linearized plasmid DNA containing a fully complementary target sequence (top) or a target sequence mismatched to the sgRNA at positions 1–2 (bottom). e, Crystal structures of dCas9-sgRNA bound to DNA substrates containing mismatches to the sgRNA at positions 1–2 (top) and 1–3 (bottom), overlaid with refined 2mFo-DFc electron density maps (grey mesh, contoured at 1σ). The sgRNA is identical to that in Fig. 1a. In both structures, the target DNA strand is provided in two fragments, as indicated in the schematics. Residual electron density corresponding to the +1 base pair is indicated with a red arrowhead.