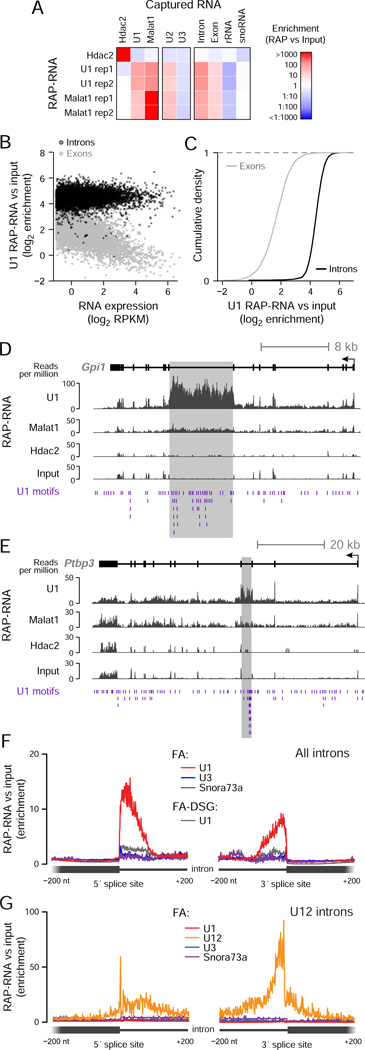
Figure 2. RAP-RNA Captures Both Direct and Indirect RNA-RNA Interactions.
(A) Enrichment for individual transcripts or classes of RNAs. RAP-RNA[FA-DSG] experiments include two replicate U1 and Malat1 purifications (rep1 and rep2) and one Hdac2 purification (negative control).
(B) Each point represents the average U1 RAP-RNA[FA-DSG] enrichment across all introns (black) or exons (gray) for one gene. The x-axis represents the expression of the mature transcript (exons) in input nuclear-enriched RNA in reads per kilobase per million (RPKM).
(C) Comparison of U1 RAP-RNA[FA-DSG] enrichment for the exons (gray) and introns (black) of all genes.
(D) RAP-RNA[FA-DSG] sequencing reads mapping to Gpi1 and (E) Ptbp3, two of the most highly enriched transcripts in U1 RAP-RNA[FA-DSG] (>100-fold versus input). Gray rectangles highlight enriched introns with many U1 motifs (purple).
(F) Enrichment of sequencing reads in RAP-RNA[FA] versus input aggregated across 5’ and 3’ splice sites for all introns (n > 180,000) or (G) U12 introns (n = 588). U1 RAP-RNA[FA] (red) more strongly enriches splice sites than U1 RAP-RNA[FA-DSG] (gray). Each read-pair contributes a count at the position corresponding to the 3’ end of the original RNA fragment for the 5’-splice-site panel and the 5’ end for the 3’-splice-site panel. See also Figure S2.