Figure 5. Factors Effecting miRNA Targeting.
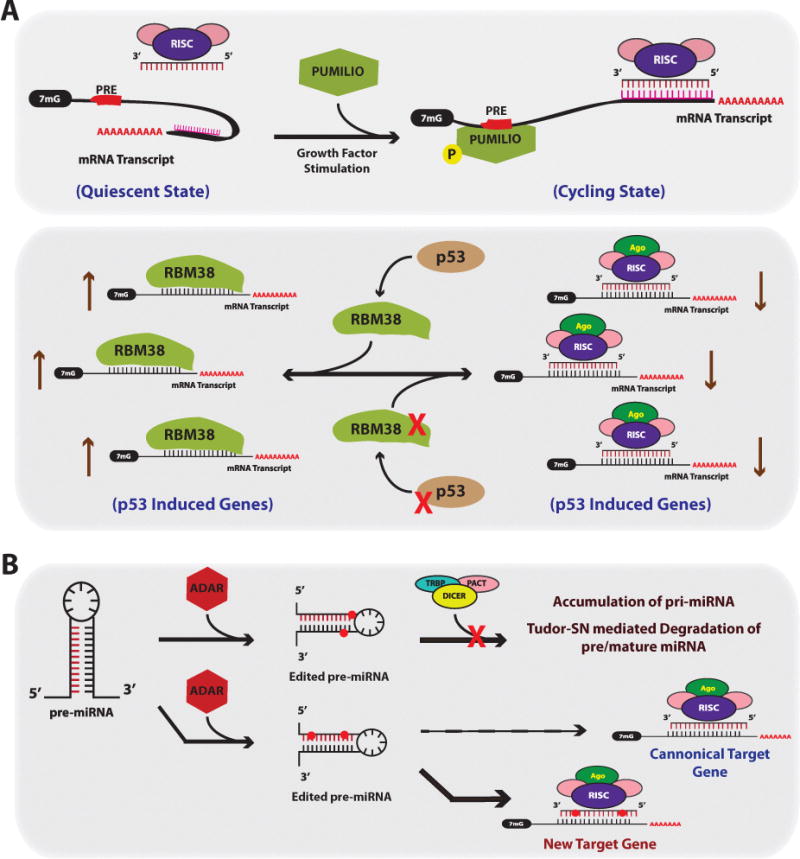
(A) RNA binding proteins can alter miRNA activity and targeting. Top Panel; certain predicted miRNA:mRNA pairing interactions may not occur under certain cell states such as quiescence, presumably due to complex or secondary structures in the mRNA preventing miRNA-RISC binding. After growth factor stimulation, transcripts that harbor PUMILIO Response Elements (PREs), will bind to PUMILIO resulting in a change in the secondary structure of the mRNA such that the predicted miRNA target site becomes accessible for RISC binding. Bottom Panel; RBM38 is an RBP that inhibits miRNA function by directly competing for RISC binding in U-rich regions in the 3′UTRs of mRNA transcripts. RBM38 is induced in a p53 dependent manner after induction of DNA damage and protects p53-induceed genes from miRNAs. In cancer, inactivating mutations in p53 and/or loss of RBM38 would result in aberrant targeting and reduction in p53 induced genes. (B) RNA editing controls both miRNA levels and proper targeting. ADAR can mediate adenosine to inosine editing of miRNA residues. Edited residues near the stem loop of the miRNA results in an unrecognized substrate for Dicer, and subsequently results in accumulation of pri-miRNA and Tudor-SN mediated miRNA degradation. Edited residues residing within the mature miRNA sequence results in new target gene recognition given the edited adenosine to inosine now functions as a guanine.
