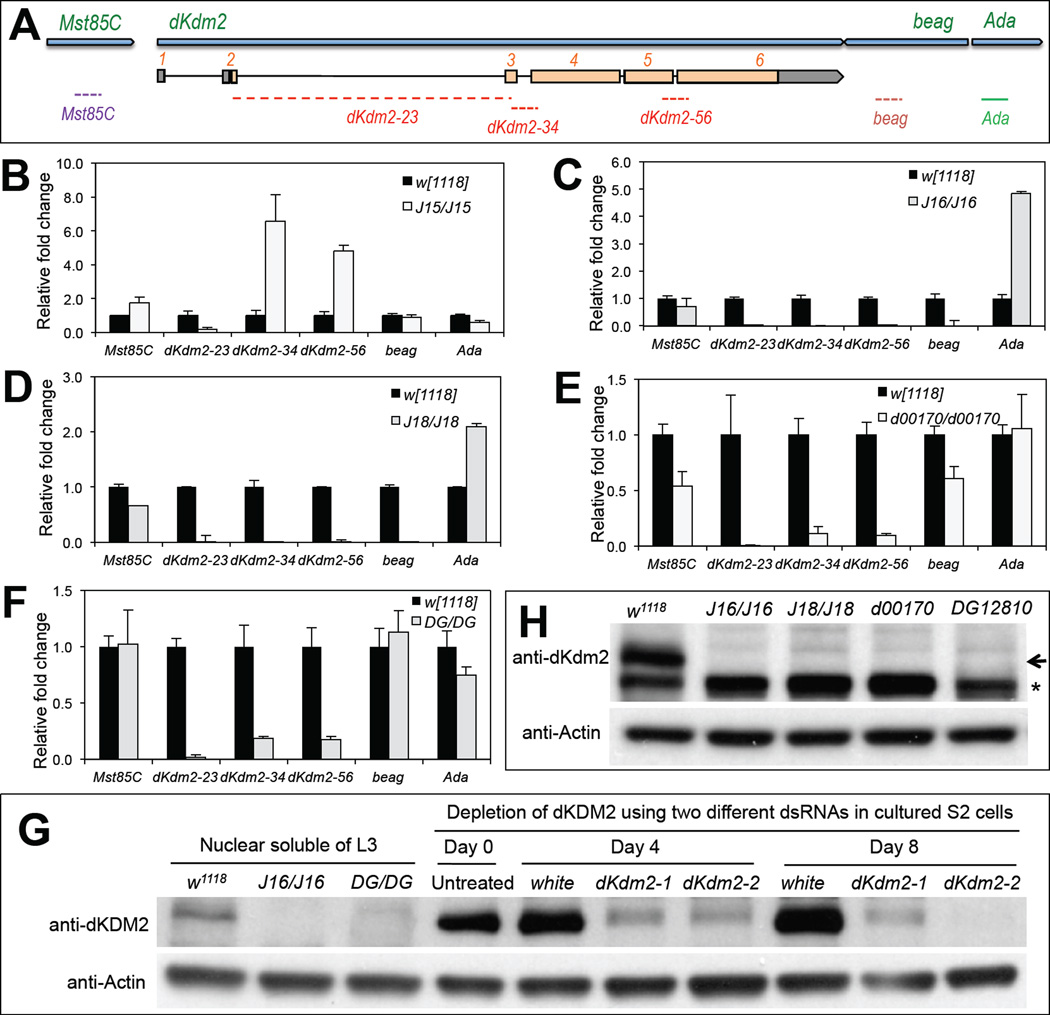
Figure 4. Effects of insertions and deletions of the class I and class II dKdm2 alleles on the expression of dKdm2 and its neighboring genes.
(A) The location of the primers used for the qRT-PCR assay. Animals of the third instar at the wandering stage were used for the qRT-PCR (C ~ F) and immunoblots (G and H). The genotypes of the mutants include: w1118; +; Df(3R)J15 (B, first instar laevar), w1118; +; Df(3R)J16 (C), w1118; +; Df(3R)J18 (D), w1118; +; dKdm2d00170 (E), and w1118; +; dKdm2DG12810 (F). (G) Western blot was used to demonstrate the specificity of the dKDM2 antiserum, note that the ~150kDa band is present in the control (w1118) but not in the Df(3R)J16 and dKdm2DG12810 mutants. In addition, this band is present in untreated S2-DRSC cells or cells treated with dsRNA to white (control), but disappears in S2-DRSC cells treated two different shRNAs targeting dKdm2. (H) Western blot of nuclear soluble proteins from the third instar larvae, showing that dDKM2 protein is undetectable in Df(3R)J16, Df(3R)J18, dKdm2d00170 or dKdm2DG12810 homozygous mutants.