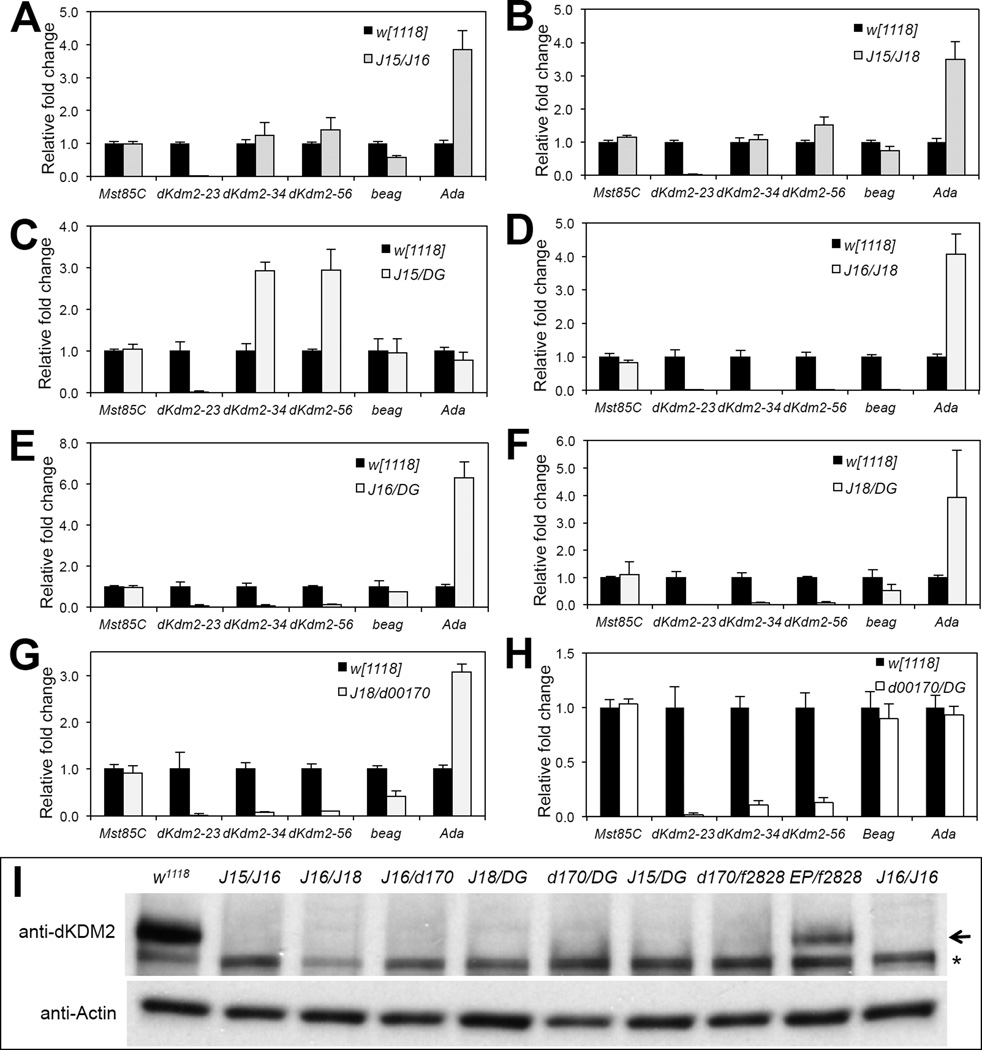
Figure 5. Expression of dKdm2 and its neighboring genes in transheterozygous combination of several class I and class II dKdm2 alleles.
All animals were collected during the third instar at the wandering stage. The genotypes include: w1118; +; Df(3R)J15/Df(3R)J16 (A), w1118; +; Df(3R)J15/Df(3R)J18 (B), w1118; +; Df(3R)J15/dKdm2DG12810 (C), Df(3R)J16/Df(3R)J18 (D), w1118; +; Df(3R)J16/dKdm2DG12810 (E), w1118; +; Df(3R)J18/dKdm2DG12810 (F), w1118; +; Df(3R)J18/dKdm2d00170 (G), and w1118; +; dKdm2d00170/dKdm2DG12810 (H). (I) Western blot of nuclear soluble proteins from these transheterozygous mutant animals at the wandering stage, note that dKDM2 protein was not detectable, except in the w1118; +; dKdm2EP3093/dKdm2f02828 mutants, which is due to the fact that the level of dKdm2 mRNA is not obviously affected in these mutants (Fig. S5E). The Df(3R)J16 homozygous mutants (denoted as “J16/J16”) was used as a control; the arrow shows the dKDM2 band, and the non-specific band is marked with a “*”.