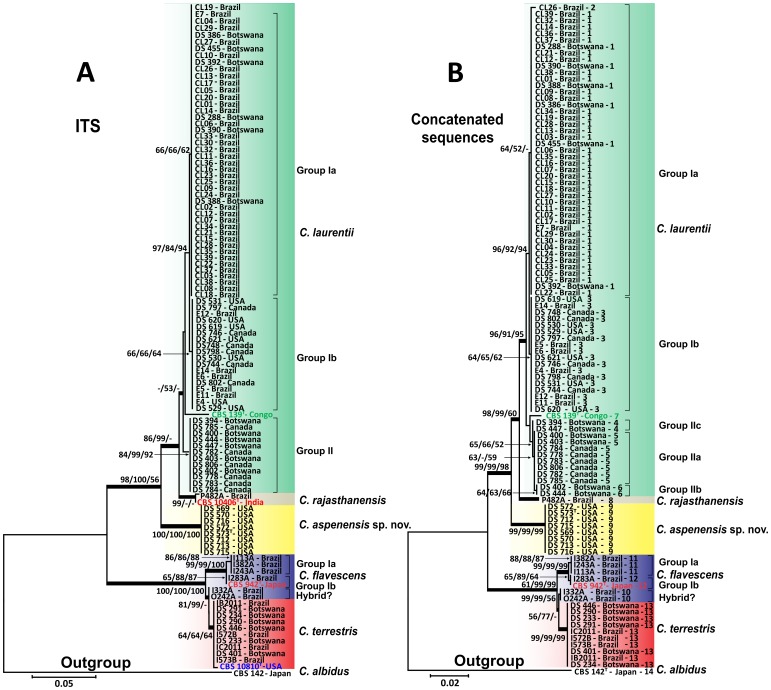
Figure 2. Phylogenetic analysis of 100 environmental Cryptococcus spp. isolates generated by the neighbor-joining, UPGMA, and maximum likelihood methods using partial nucleotide sequences of the (A) internal transcribed spacer (ITS) and (B) concatenated sequences of the three ribosomal regions.
Numbers at each branch indicate bootstrap values>50% based on 1,000 replicates (NJ/UPGMA/ML). The analysis involved 105 and 103 nucleotide sequences for ITS and concatenated sequences respectively. T: Type strain.