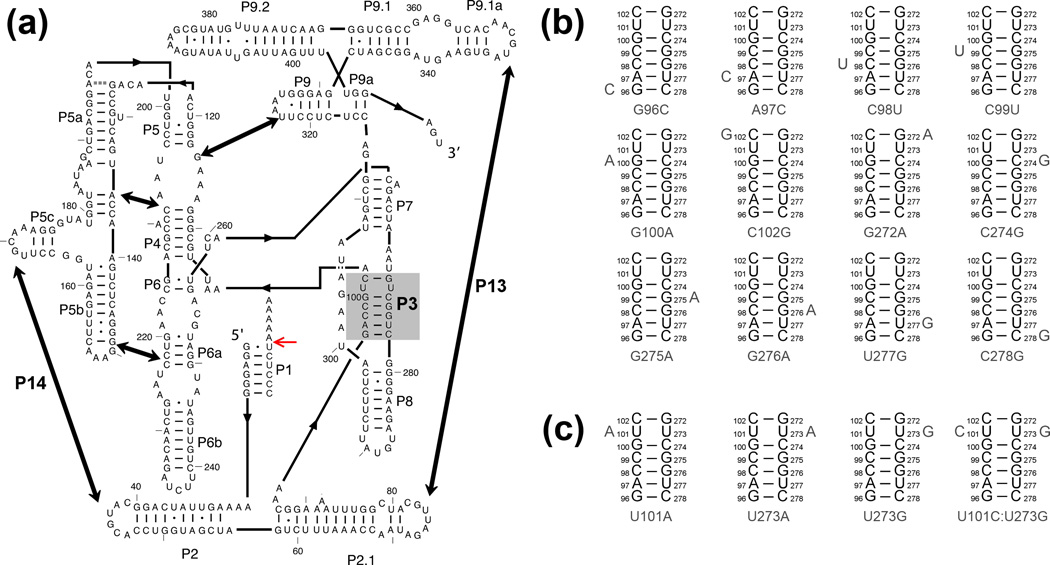
Figure 1.
The Tetrahymena ribozyme and mutants used herein. (a) Ribozyme secondary structure. The ribozyme contains five long-range contacts, as indicated by thick arrows. The ribozyme is shown with substrate bound to form the P1 helix, and the red arrow indicates the substrate cleavage site. The P3 helix is shaded in gray. (b–c) Point mutations are designed to weaken the P3 helix by disrupting a single base pair (panel b) or to strengthen P3 by creating A–U, U-A, or U-G pairs or a C–G pair (panel c).