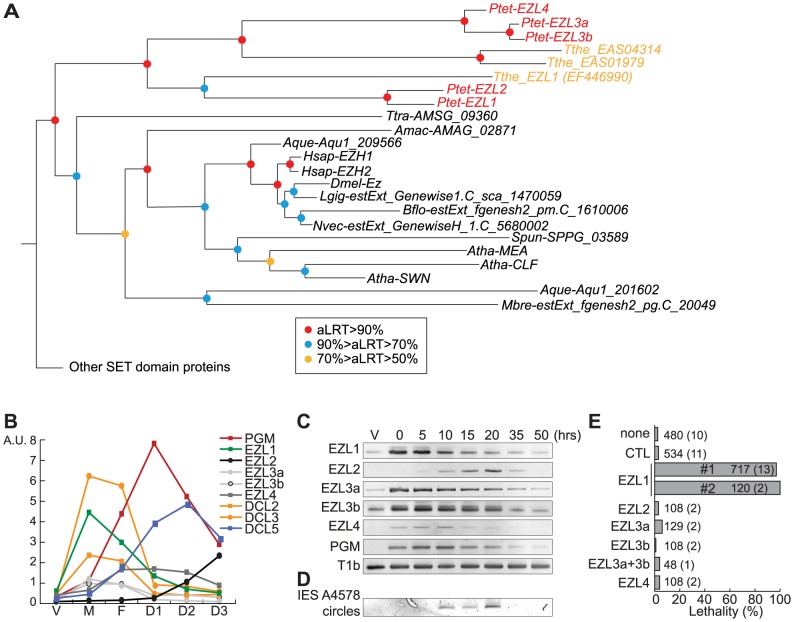
Figure 3. Identification and functional analysis of EZL genes.
A. Phylogenetic analysis of the EZH/EZL SET domain proteins. The part of the Maximum-likelihood (ML) tree shown in Figure S5 and which includes the EZH/EZL proteins, is depicted. Statistical supports (aLRT values) are indicated on the nodes by colored circles (color code is indicated in the figure). Species abbreviations: Amac = Allomyces macrogynus (Fungi); Atha = Arabidopsis thaliana (Virdiplantae); Aque = Amphimedon queenslandica (Metazoa); Bflo = Branchiostoma floridae (Metazoa); Dmel = Drosophila melanogaster (Metazoa); Hsap = Homo sapiens (Metazoa); Mbre = Monosiga brevicolis (choanoflagellata); Nvec = Nematostella (Metazoa); Ptet = Paramecium tetraurelia (Ciliata); Spun = Spizellomyces punctatus (Fungi); Tthe = Tetrahymena thermophila (Ciliata); Ttra = Thecamonas trahens (Apusozoa). B. Expression patterns of EZL genes during the life cycle. EZL, DCL2, DCL3, DCL5 and PGM gene expression levels, as determined by microarray expression data during autogamy time-course experiments [26]. The vegetative time point (V) consists of 4 samples from mass cultures containing only log-phase cells showing no sign of meiosis. The meiosis time point (M) consists of 4 samples containing 20-39% of cells undergoing meiosis, and little or no fragmentation of the maternal MAC. The fragmentation (F) time point consists of 4 samples that contained a similar proportion of meiotic cells (20-29%) as the M time point, but also contained 37-43% of cells with a fragmented maternal MAC. The D1 time point groups 3 samples with 35-56% of cells with fragmented maternal MACs and 35-51% of cells that already contained clearly visible new MACs. D2 consists of 3 samples with 73-98% of cells with visible new MACs, and the D3 samples were taken ∼10 hours after the D2 samples. C. Detection of EZL and PGM mRNA during autogamy by RT-PCR. Total RNAs were extracted at each time point (see Fig. S7), were reverse transcribed and cDNAs were amplified by PCR with gene specific primers and, as a loading control, with primers for the T1b gene, which encodes a component of the secretory granules [55]. D. PCR detection of IES 51A4578 circles with divergent primers on genomic DNA at each time point shown in Figure S7 after ICL7 (control) or EZL1 silencing. E. Lethality of post-autogamous progeny following EZL gene silencing. The gene targeted in each silencing experiment is indicated. Two non-overlapping fragments (#1 and #2) of EZL1 gene were used independently. The ND7 or ICL7 genes were used as control (CTL) RNAi targets, since their silencing has no effect on sexual processes [27]. Autogamy was also performed in standard K. pneumoniae medium (none). Cells were starved in each medium to induce autogamy and, following 3-4 days of starvation, autogamous cells were transferred individually to K. pneumoniae medium to monitor growth of sexual progeny. The total number of autogamous cells analyzed for each RNAi and the number of independent experiments (in parenthesis) are indicated. Death in progeny after EZL1 silencing was observed after less than three cell divisions. The absence of lethality observed after EZL2, EZL3a, EZL3b, EZL4 KDs should be taken with caution as the level of KDs was not measured.