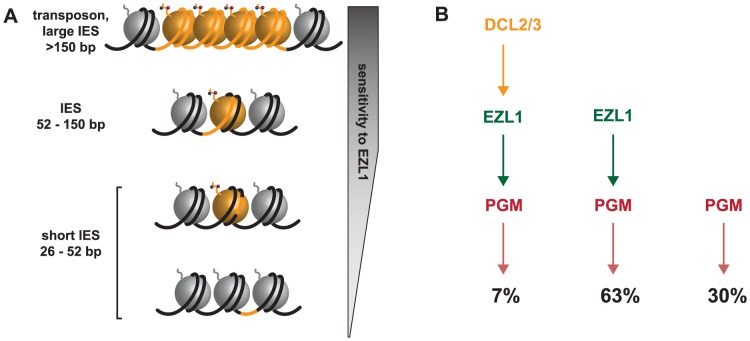
Figure 7. Model for the action of the histone methyltransferase Ezl1 in programmed genome rearrangements and schematic representation of partially overlapping pathways involved in IES excision.
A. To take into account the length bias of DNA sequences retained after Ezl1 depletion, we propose that efficient excision of germline-limited DNA segments (orange) is regulated by the position of methylated nucleosomes. For long DNA segments (>150 bp in length) that are covered by at least one nucleosome, the putative HMT Ezl1 would be targeted to the eliminated sequences and would catalyze the trimethylation of H3 K27 and K9. These histone marks would attract, or activate, the excision machinery, thereby the excision of marked DNA segments. DNA segments whose size is comprised between 52 bp and 150 bp would be partly or entirely included within one nucleosome and histone H3 methylation would be essential for their recognition and excision. The smallest DNA segments (26-52 bp in length), however, would be either wrapped around a nucleosome and histone methylation might be needed for efficient excision, or located within the linker DNA and histone methylation might be dispensable for their excision. B. Schematic representation of partially overlapping pathways involved in IES excision. While all IESs are ultimately excised by the Pgm endonuclease, there appears to be different classes of IESs. Some IESs (30%) require neither EZL1 nor DCL2/3 for complete excision; others (7%) require EZL1 and DCL2/3; while the majority require only EZL1 (66%).