Figure 3. SAM-I superfamily.
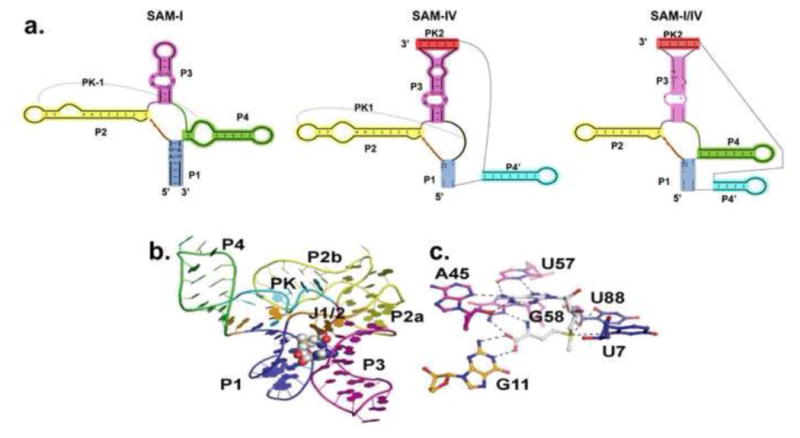
a.) Side-by-side comparison of the secondary structures of SAM-I, SAM-IV, and SAM-I/IV riboswitch families[10]. Equivalent helical elements among three families are drawn in the same color. Pseudoknot-1 (PK-1) structure is marked by a dashed line. b.) Representative SAM-I crystal structure from T. tengcongensis[25]. The RNA is in cartoon representation and colored according to panel a. SAM is in CPK representation. c.) Zoomed view of the SAM binding site in SAM-I riboswitch. SAM ( in white color) forms a base triple with A45 and U57. G58 and G11 interact with the methionine tail of SAM.
