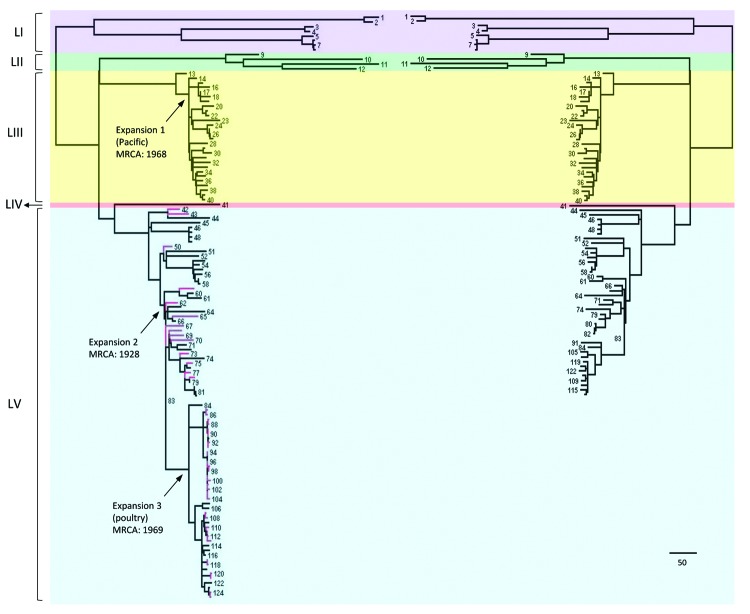
Figure 1.
Comparison of Salmonella enterica serotype Enteritidis phylogenies inferred from Illumina data and combined data of Illumina (San Diego, CA, USA) and Roche 454 (Indianapolis, IN, USA). The tree on the right incudes 80 Illumina sequenced isolates and the reference genome (PT4). The tree on the left includes both the 80 Illumina and the 44 454 sequenced isolates in addition to the reference. Isolates were numbered (online Technical Appendix Table 1, http://wwwnc.cdc.gov/EID/article/20/9/13-1095-Techapp-s1.pdf). Lineages I, II, III, IV, and V are highlighted in purple, green, yellow, red, and blue, respectively. Branches representing 454 sequenced isolates are labeled in red. Arrows on the left tree indicate the 3 serotype Nitra isolates. MRCA, most recent common ancestor. Scale bar indicates 10 single-nucleotide polymorphisms.