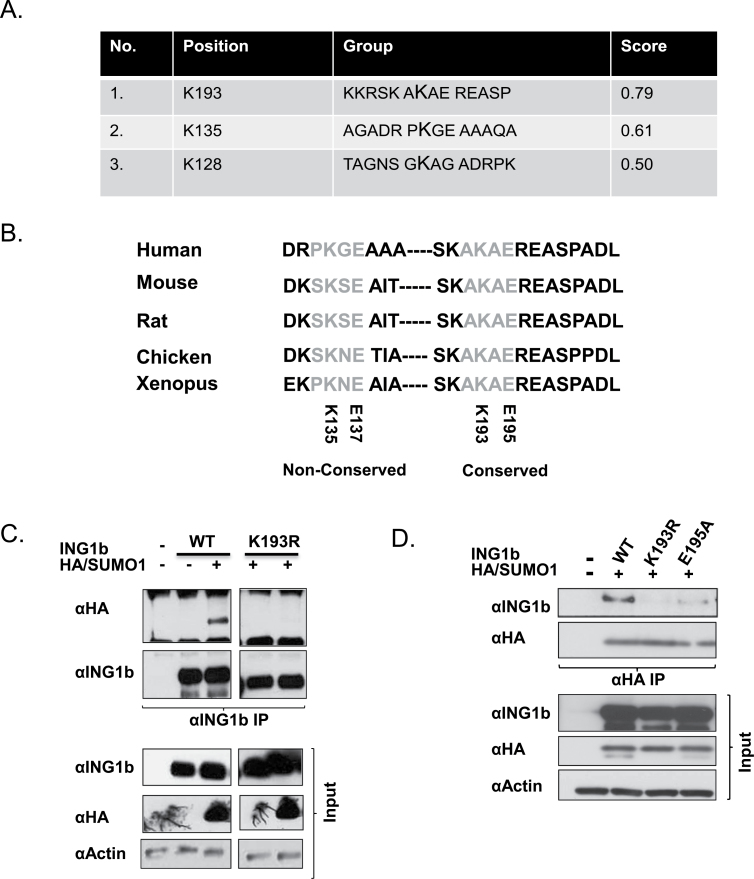
Fig. 3.
Identification of ING1b SUMO acceptor sites. (A) SUMOplot software (Abgent) predicts the presence of three consensuses SUMO sites from the primary protein sequence of ING1b and a score is assigned based on the hydrophobicity of the first residue and the presence of an acidic amino acid on the fourth position of the consensus motif. (B) Sequence analysis revealed that the site with the highest probability containing lysine 193 is completely conserved in other vertebrates. (C) αING1b denaturing IP was performed with HEK293 cell lysates expressing ING1b wild-type (WT) and the ING1 K193R (K193R) point mutant with and without HA/SUMO1. Expression of the transfected constructs and equal loading were confirmed using αING1 and αHA (SUMO1) and α-actin, respectively. (D) A reciprocal αHA IP was performed with U2OS cell lysates expressing ING1b WT, K193R and E195A (SUMOylation specific point mutant) and HA/SUMO1 to confirm the presence of a functional SUMO consensus site (AKAE) containing lysine 193 and glutamic acid 195.