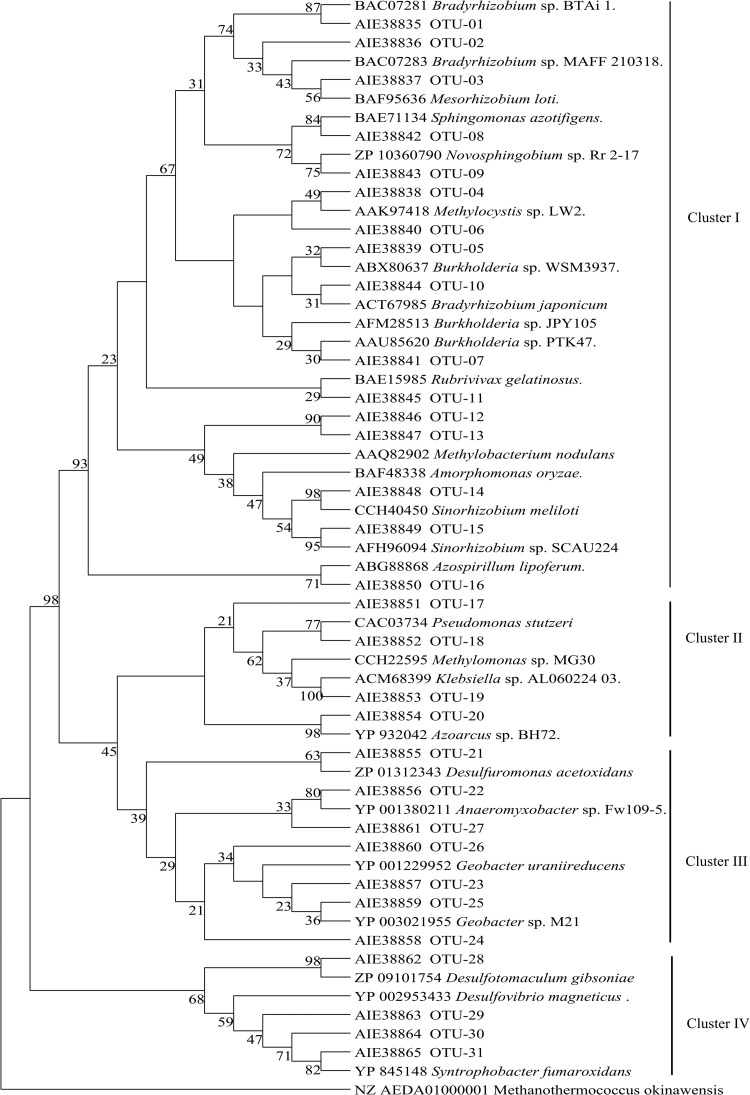
FIG 3.
Phylogeny and composition of nifH phylotypes representing four or more sequences. OTUs from this study are represented by OTU number and the associated protein accession number. The evolutionary history was inferred by using the maximum likelihood method based on the Poisson correction model. The bootstrap consensus tree was inferred from 1,000 replicates. There were a total of 113 positions in the final data set, and analyses were conducted in MEGA5. Sequences from known bacteria are indicated by name and NCBI protein accession numbers. This tree was rooted with the nifH gene from the archaeon Methanothermococcus okinawensis.