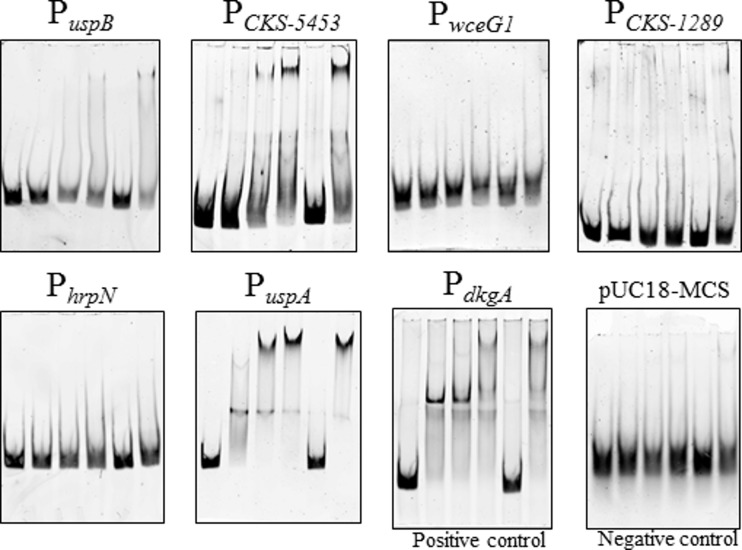
FIG 2.
EMSAs of promoters identified using bioinformatics tools. Promoters of potential direct targets of EsaR identified via a primarily bioinformatics approach were analyzed for binding by the EsaR fusion protein, HMGE. The promoter of dkgA (13) was used as a positive control, and a 32-bp pUC18 MCS DNA fragment was used as a negative control. The concentration of FAM-labeled DNA probe in all lanes is 5 nM. The lanes within each panel consist of the following (left to right): DNA probe, DNA probe with 100 nM HMGE, DNA probe with 200 nM HMGE, DNA probe with 400 nM HMGE, DNA probe with 400 nM HMGE and 1 μM specific competitor PesaR28 DNA fragment, and DNA probe with 400 nM HMGE and 1 μM nonspecific pUC18 MCS DNA fragment.