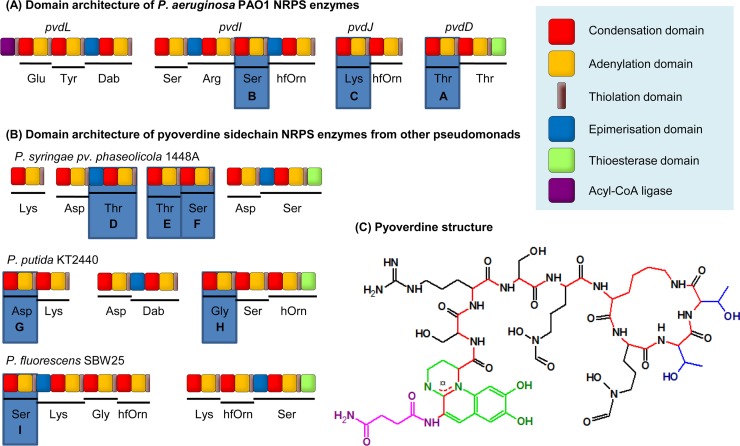
FIG 2.
NRPS enzymes, labeled from A to I, with modules used for domain substitutions highlighted. (A) Domain architecture of the four NRPS enzymes involved in P. aeruginosa PAO1 pyoverdine synthesis, based on previous annotation (15). (B) Domain architecture of the NRPS enzymes involved in synthesis of the pyoverdine variable peptide chains from three other Pseudomonas strains. An additional NRPS, PvdL (as shown in panel A), is conserved in each strain and incorporates the residues that later form the chromophore. The figure is based on previous annotation by Owen and Ackerley (40) (P. syringae 1448a), Moon et al. (41) (P. fluorescens SBW25), and Ravel and Cornelis (42; adapted with permission of the publisher) (P. putida KT2440). (C) Structure of pyoverdine from P. aeruginosa PAO1 (43, 44) with the succinimide group highlighted in pink, the chromophore in green, the peptide backbone in red, and the side chains of the threonine residues incorporated by PvdD in blue. Dab, 2,4-diaminobutyric acid; hfOrn, l-N5-formyl-N5-hydroxyornithine; hOrn, N5-hydroxyornithine.