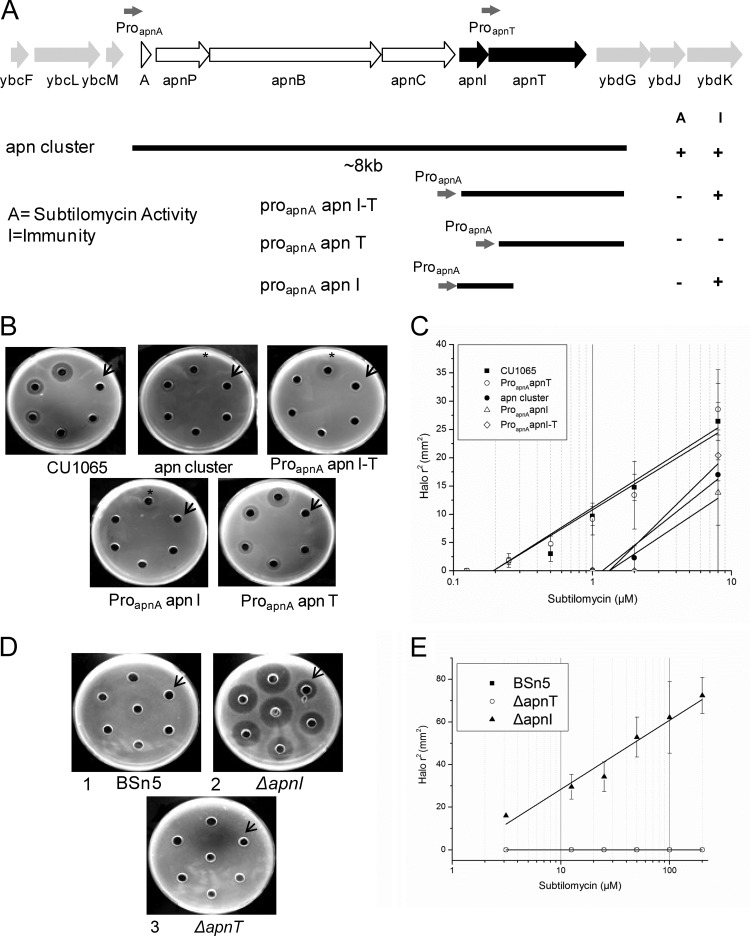
FIG 4.
Functional analysis of subtilomycin immunity by gene expression in B. subtilis CU1065 and gene interruption in B. subtilis BSn5. (A) Map displaying essential DNA fragments for subtilomycin immunity. Each construction was cloned into the vector pDG1730 and integrated into the amyE locus of the chromosome of the subtilomycin-sensitive strain B. subtilis CU1065. The predicted fragments containing the apnA and apnT promoter regions are indicated by gray arrows. (B) Subtilomycin sensitivities of B. subtilis CU1065 strains harboring different fragments of the subtilomycin gene cluster were investigated in agar diffusion tests. B. subtilis CU1065 and its recombinants were transformed with the vector containing proapnA-apnI-T, proapnA-apnI, proapnA-apnT, or the whole subcluster. Starting from the arrow and moving clockwise, the subtilomycin concentrations applied were as follows: 0.125 μM, 0.25 μM, 0.5 μM, 1 μM, 2 μM, and 8 μM. The asterisk indicates a blurry inhibition zone. (C) Linear dependencies between the square of the radius of the halos shown in panel B and the natural logarithm of the applied subtilomycin concentration with 60 μl of subtilomycin and diffusion for at least 4 h. Results for B. subtilis CU1065 transformed with vectors containing apnI, apnT, apnI-apnT, and subgene clusters are shown. (D) Subtilomycin immunity of strain BSn5 (plate 1) with interruption of apnI (plate 2) and apnT (plate 3). Starting from the arrow and moving clockwise, the applied concentrations of subtilomycin were 3.125 μM, 12.5 μM, 25 μM, 50 μM, 100 μM, and 200 μM. The well in the middle of the plate was not counted. (E) Subtilomycin immunity of BSn5 with interruption of apnT and apnI. The immunity response was calculated corresponding to the square of the radius of the halos shown in panel D.