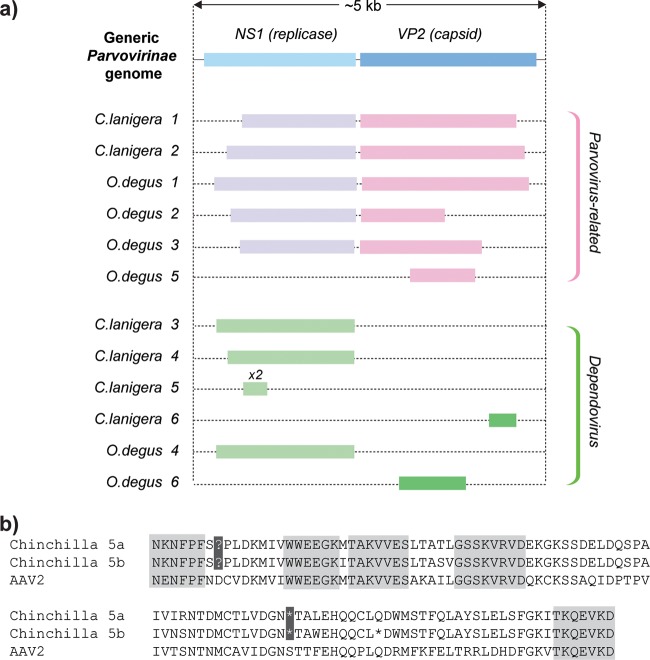
FIG 1.
(a) Genetic structures of parvovirus-derived EVEs in C. lanigera and O. degus genomes. Open reading frames (ORFs) were inferred by manual comparison of putative peptide sequences to those of closely related exogenous parvoviruses. (b) Alignment of a pair of duplicated EVEs in the Chinchilla lanigera genome against an adeno-associated virus type 2 (AAV2) reference sequence (GenBank accession no. AF043303.1). Blocks of six or more amino acids that are conserved between all three sequences are highlighted in gray. Nonsense mutations shared between the two duplicated EVEs are highlighted in black. The two EVEs differed at 8 of 323 nucleotide positions (i.e., an average of 4 nucleotide substitutions have occurred in each insertion), indicating that these EVEs arose in a duplication event that occurred between 2.8 and 5.6 MYA (assuming a range of mammalian neutral substitution rates from 2.2 × 109 to 4.5 × 109 per site per year [17]).