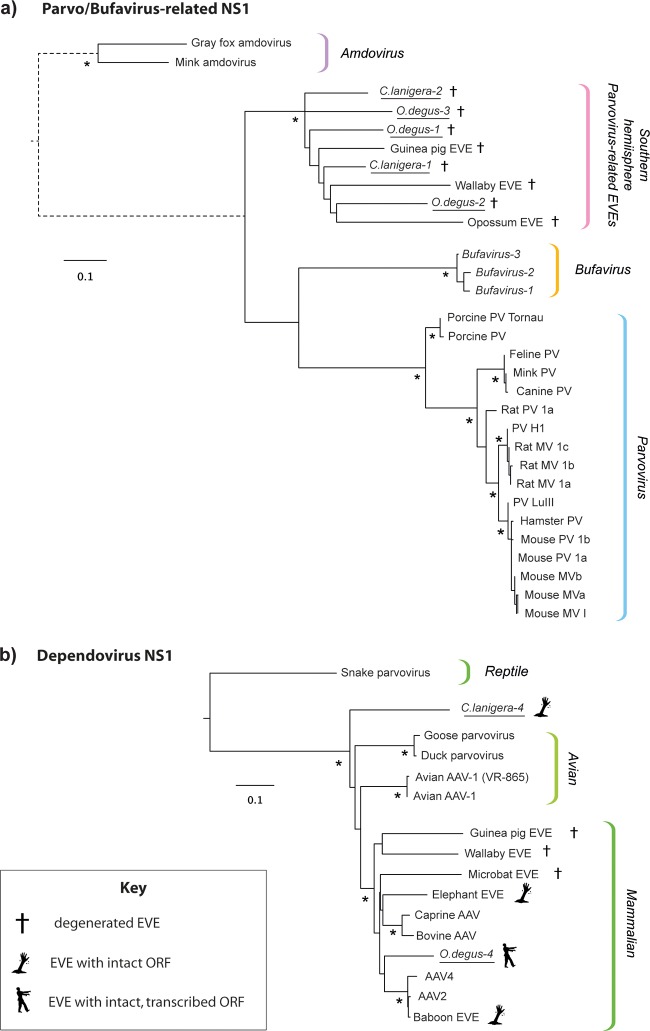
FIG 2.
Maximum likelihood phylogenies showing the relationships of parvovirus-related (a) and dependovirus-related (b) EVEs in the C. lanigera and O. degus genomes to exogenous parvoviruses and previously described EVEs. Phylogenies are based on alignments of NS1 proteins and putative NS1 proteins encoded by EVE pseudogenes and were constructed using PHYML (18) and the JTT+ protein substitution model as selected by ProtTest (19). EVEs identified in this study are underlined. Phylogenies are midpoint rooted for clarity of presentation. The scale bar indicates evolutionary distance in numbers of substitutions per amino acid site. Branches shown as dashed lines have been shortened and are not shown to scale. Asterisks indicate nodes with maximum likelihood bootstrap support levels above 75% for 100 bootstrap replicates. EVE, endogenous viral element; AAV, adeno-associated virus; MV, minute virus; PV, parvovirus; NS, nonstructural protein gene cassette; VP, viral capsid protein gene cassette.