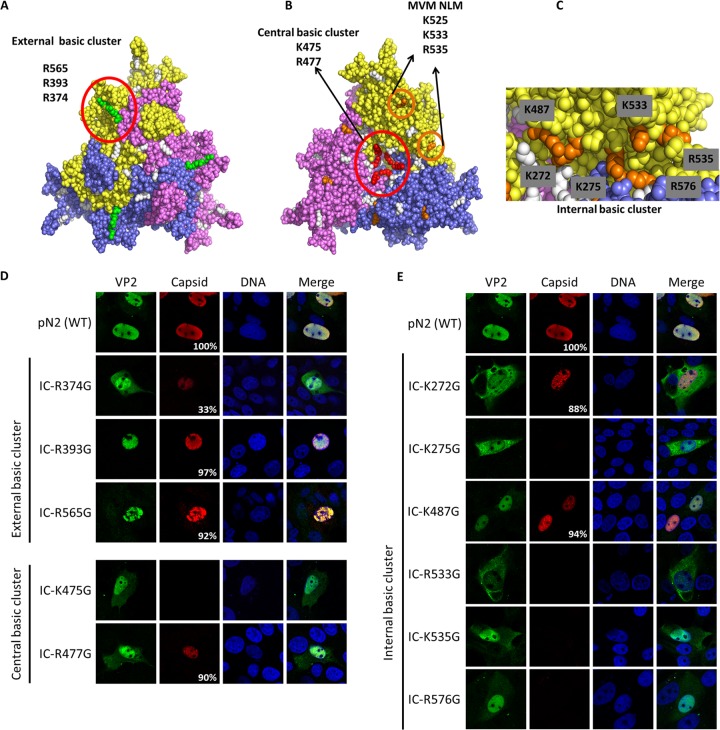
FIG 5.
NLM identification for VP2 protein. Three basic clusters were identified using the trimer structure information (Protein Data Bank accession number 1K3V) (basic amino acids are in white). (A) One external basic cluster is located outside the capsid and includes three basic amino acids (green). (B) A central basic cluster is located inside the capsid at the center of the trimer and includes two basic amino acids for each protein (a total of 6 amino acids; red). The image also shows basic amino acids in the beta-sheet corresponding to MVM NLM (orange). (C) Another internal cluster is located inside the capsid and is partly conserved with the NLM of MVM and includes six basic amino acids (orange). Basic amino acids were replaced by glycines in the infectious clone of PPV (IC). VP2 localization and capsid formation were investigated in transfection experiments. Immunofluorescence experiments were performed with rabbit anti-VP2 antibody together with anti-rabbit Alexa Fluor 488 antibody and mouse anti-capsid antibody together with anti-mouse Alexa Fluor 568 antibody. (D) Mutations in the external basic cluster or central basic cluster did not significantly change global VP2 localization. (E) Mutation of some of the basic amino acids of the internal cluster resulted in a VP2 located either in a diffuse pattern between the nucleus and the cytoplasm or mostly in the cytoplasm. All images are representative fields for each clone from at least three transfections. For each clone at least 300 positive cells were analyzed. The percentage of cells with capsid formation is also indicated.