FIG 8.
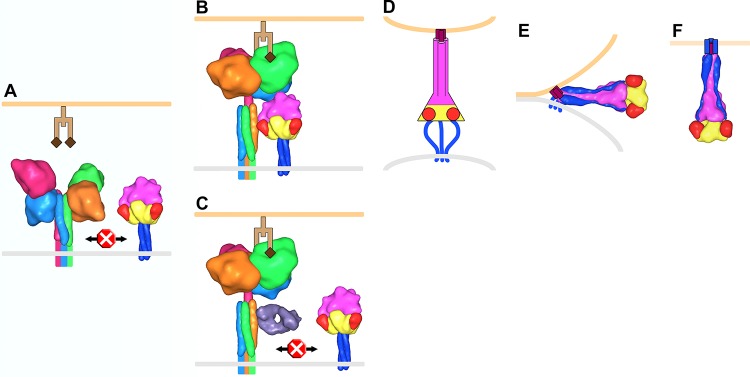
Model of PIV5 membrane fusion. (A) F trimers (yellow, red, magenta, and blue corresponding to domains I, II, III, and HRB, respectively) and HN tetramers (monomers shown as light blue, hot pink, orange, and green) reside on the surface of the virus membrane (gray). Structural data are lacking for the membrane-proximal portion of the HN stalk, and this region, the transmembrane domain, and the cytoplasmic tails are depicted as colored lines. Prior to receptor (light and dark brown representing a glycan chain terminating with sialic acid) engagement on a host cell membrane (light orange), HN is in a four-heads-down conformation in which the heads sterically block interaction between the membrane-distal portion of the HN stalk and the head domain of F. (B) HN head domains transition to a four-heads-up position upon receptor engagement allowing the F/HN interaction. HN is rotated ∼45° clockwise in panels B and C compared to the orientation in panel A. (C) Antibodies (Fab, violet) that bind to the membrane-distal portion of the HN stalk inhibit membrane fusion by blocking the interaction with F. (D) The F-HN interaction results in a major conformation change in F such that domain III refolds into an extended trimeric coiled coil (HRA) and inserts a hydrophobic fusion peptide (purple) into the target cell membrane (for simplicity HN and receptor are not shown in panels D to F). Structural data are lacking for this “prehairpin intermediate” conformation; therefore, it is depicted exclusively as a cartoon. (E) HRB interacts with HRA, forming a thermostable six-helix bundle domain in the postfusion conformation, which juxtaposes the virus and target cell membranes leading to membrane fusion (F). To make this figure, Sculptor (version 2.1.1_r1 [http://sculptor.biomachina.org]) (69) was used to morph high-resolution structures of HN, F, and the humanized anti-HER2 Fab, 4D5 (which the sAb phage library was based on), to 15-Å surface representations. Protein Data Bank (PDB) entries 3T1E and 3TSI were overlaid and used for the HN four-heads-down model. Similarly, PDB 1Z4X, 3TSI, and 4JF7 were used for the HN four-heads-up model. PDB entries 2B9B and 1ZTM were used for the pre- and postfusion representations of F, respectively. The GCNt trimerization domain was used as a surrogate to represent the TM and CT domains of prefusion F. PDB 1FVD was used for the Fab.
