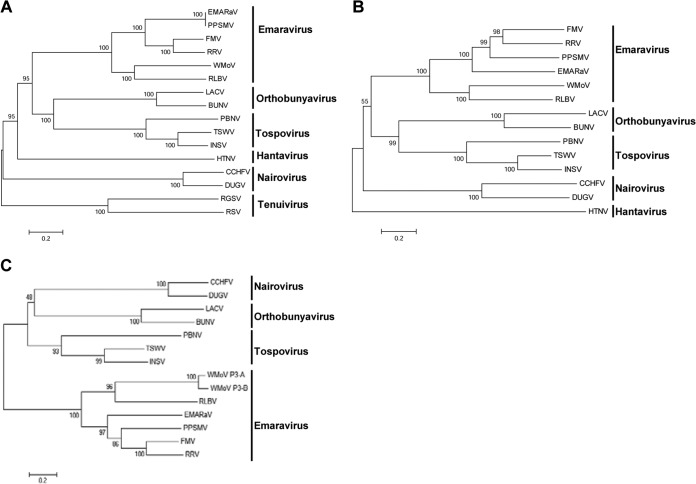
FIG 5.
Phylogenetic analyses of emaraviruses, representative members of Bunyaviridae, and Tenuivirus. Unrooted bootstrap consensus phylogenetic trees were generated from the amino acid sequences of RdRp (A), glycoprotein precursor protein (B), and nucleocapsid protein (C). Phylogenetic trees were constructed by the neighbor-joining method using the JTT matrix and pairwise gap deletion with 1,000 bootstrap replicates; bootstrap support is indicated at branch points. The bar represents the number of amino acid replacements per site. Note that WMoV formed a separate clade with RLBV from other members of the genus Emaravirus. GenBank accession numbers of proteins and the names of viruses used for phylogenetic analyses are given in Tables 2 and 3.