FIG 4.
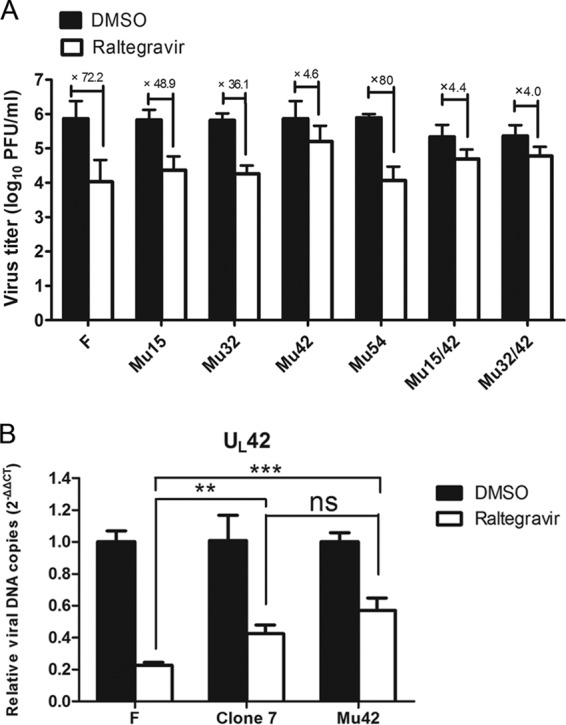
A single amino acid mutation in UL42 confers raltegravir resistance. (A) Identification of the virus titers of the recombinant viruses corresponds to different amino acid mutations. Subconfluent CV1 cells in 6-well plates were infected with recombinant viruses as well as HSV-1(F) at an MOI of 0.01 PFU per cell. After 60 min of incubation at 37°C, the inocula were removed, residual extracellular infectivity associated with the cells was reduced by treatment with a low-pH citrate buffer, and growth media containing 200 μM raltegravir were added. At 24 hpi, the amount of infectious virus was determined by plaque assay in CV1 cells. (B) Comparison of the viral DNA copies of HSV-1(F), clone 7, and Mu42 with DMSO treatment or raltegravir treatment using real-time qPCR. CV1 cells were infected with these three viruses at an MOI of 5 PFU per cell. After virus adsorption and washing, cells were maintained in medium containing 200 μM raltegravir or an equivalent of DMSO carrier. The cells were lysed, and total cellular DNA was extracted and used as the template for qPCR, as described previously (see Fig. 2C). qPCR was performed in triplicate, and data are shown as the mean ± SD of three independent experiments. ns, not significant (P > 0.05); **, P < 0.01; ***, P < 0.001.
