FIG 4.
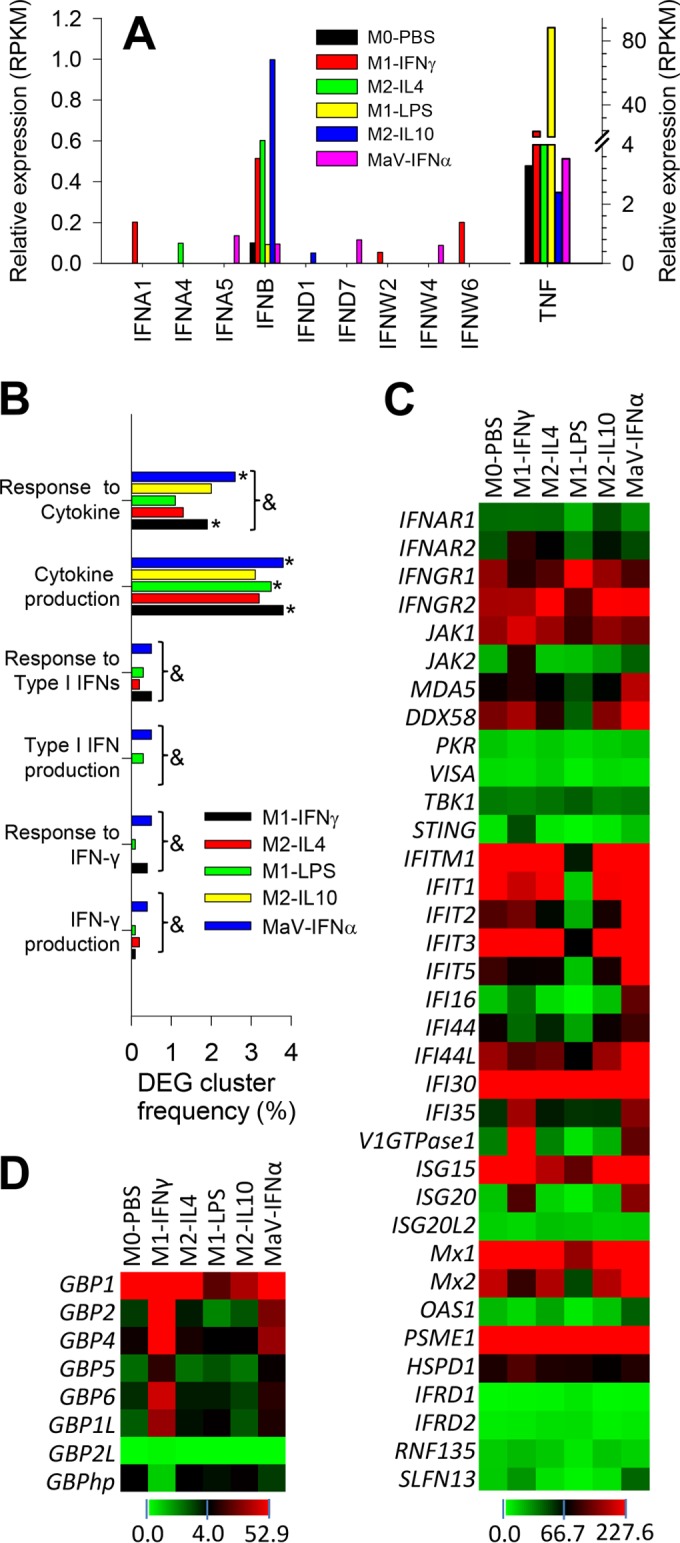
Quantitative expression and Gene Ontology (GO) analyses of interferon (IFN) production and action. (A) Quantitative expression analysis of type I IFN transcripts in PRRSV-infected macrophages at different activation statuses, showing suppressed type I IFN expression compared with the detection of TNF (tumor necrosis factor alpha). (B) GO analysis of DEGs clustered with IFN production and action compared with DEG cluster frequency to other cytokines, * and &, P < 0.05 compared with M0-PBS or among all statuses, respectively. (C) Heat map of the differential expression of genes encoding IFN receptors and IFN-stimulated/induced genes (ISGs/IFIs). (D) Family-wide analysis of the expression of genes encoding guanylate binding proteins (GBPs), a group of ISGs recently shown to be potentially associated with PRRSV pathogenesis (44). GBPhp is an unknown hypothetical gene that formed into a gene cluster with all other porcine GBP genes in chromosome 4 and had an expression pattern negatively correlated with most other porcine GBP genes. The color scale under each heat map illustrates the midpoint and range of RPKM values of the listed transcripts.
