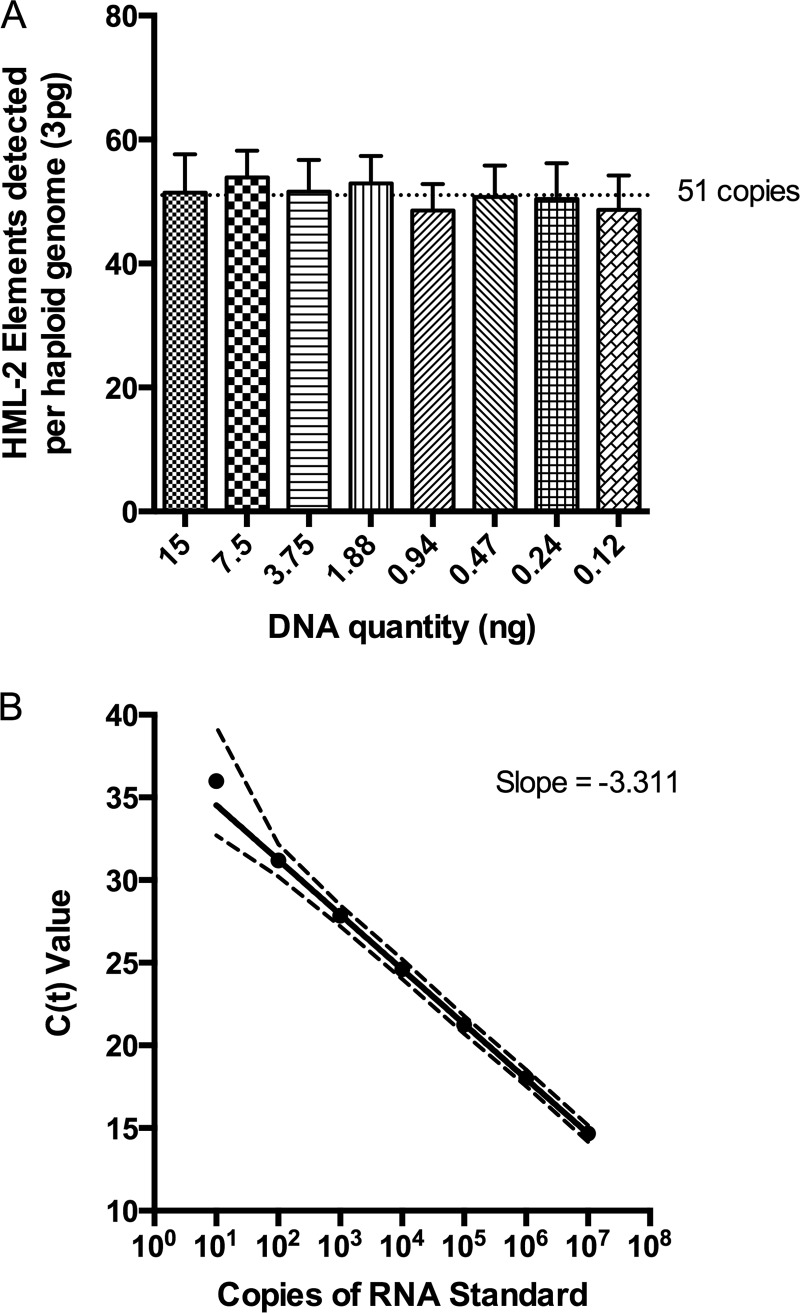
FIG 1.
Quantitative-PCR detection of HML-2 proviruses. (A) The number of HML-2 proviruses detected per haploid genome using env qPCR was determined using dilutions of human genomic DNA at known concentrations (using the estimate of 6 pg DNA/cell and 3 pg DNA/haploid genome) and determining the copy numbers of HML-2 DNA at these different dilutions using plasmid DNA standards containing the 7p22.1a env sequence. The mean and standard deviation are plotted for each dilution, where the mean is the average of replicate wells from 3 assays. (B) RNA standard serial dilutions were reverse transcribed in duplicate, and the cDNA was assayed using SYBR green env qPCR. Mean cycle threshold values (CT; y axis) and the 95% confidence intervals (CI) are plotted for each dilution of the RNA standard (x axis). CT values are compiled from multiple assays (n = 41 for 10 to 106 copies and n = 26 for 107 copies). The slope of the line is −3.311, corresponding to a PCR efficiency of 100.46%.