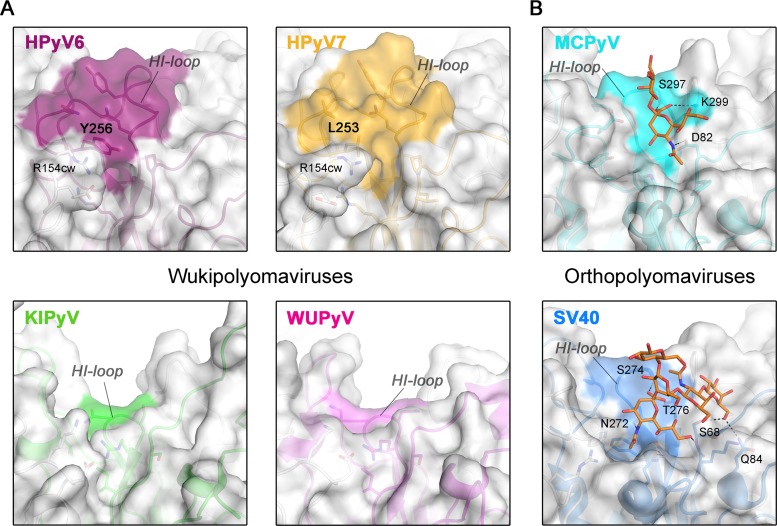
FIG 3.
Surface structures of VP1 pentamers. Closeup views of VP1 pentamer top-surface regions that are involved in sialic acid engagement in the case of MCPyV and SV40 are shown. Equivalent surface sections are shown in surface and cartoon representations for VP1 pentamers from HPyV6, HPyV7, and KIPyV (PDB accession no. 3S7V) and WUPyV (PDB accession no. 3S7X) (A) and from SV40 (PDB accession no. 3BWR) and MCPyV (PDB accession no. 4FMI) (B). HI-loop residues are highlighted on the surface representations according to the colors assigned to the respective viruses. Carbohydrates (Neu5Ac and Gal of α2,3-sialyllactosamine and GM1 pentasaccharide) in panel B are shown in stick representations (colored by atom type; carbons in orange, oxygen in red, and nitrogen in blue), and glycan-protein contacts (hydrogen bonding and salt bridges) are shown as dashed lines for MCPyV and SV40 VP1.