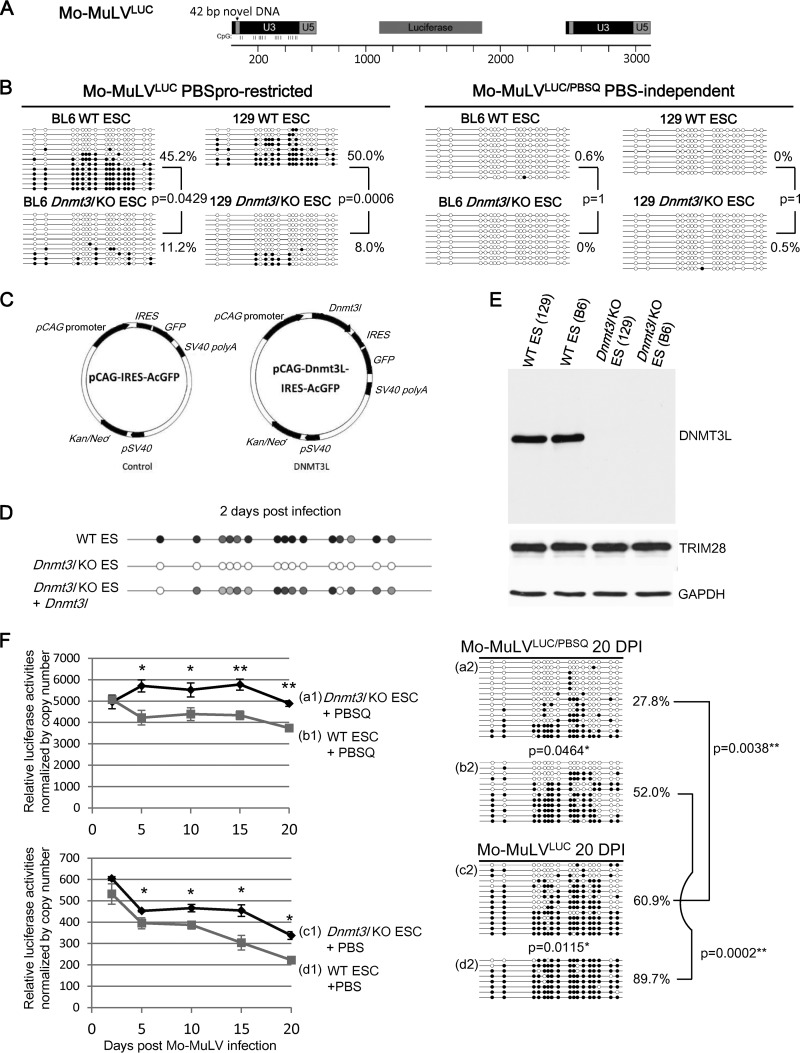
FIG 1.
DNMT3L and the ZFP809-TRIM28 pathway are both required for epigenetic silencing of Mo-MuLV in ES cells. (A) Retroviral Mo-MuLV construct used in all studies containing a luciferase gene and a 42-bp insert within the U3 region of the Mo-MuLV long terminal repeat (LTR). This insert provides specific detection against preexisting and endogenous Mo-MuLV elements. The proviral DNA CpG sites for bisulfite sequencing are indicated. (B) ES cells lacking DNMT3L cannot add methylation marks to the LTR of proviral Mo-MuLV at 2 days postinfection. The PBSpro sequence from Mo-MuLV is also required for efficient de novo DNA methylation. DNA from wild-type (WT) or Dnmt3L KO ES cells was isolated at 2 days after Mo-MuLVLUC or Mo-MuLVLUC/PBSQ infection and analyzed by bisulfite sequencing using primers specific for the 42-bp insertion that were added to the LTRs of the Mo-MuLV constructs. The presence of a methylated or nonmethylated CpG within each sequence is indicated using a black or white circle, respectively. B6, C57BL/6 mice; 129, 129S4/SvJae mice. (C) Control and Dnmt3l expression vectors containing a neomycin resistance gene and IRES-GFP driven by a pCAG promoter. (D) Transfection of a Dnmt3l-expression vector rescues the methylation level in Dnmt3l KO ES cells. The bisulfite PCR product sequencing results are presented. The gray circles indicate the partially methylated CpG sites of mixed PCR fragments. (E) Protein expression levels of DNMT3L, TRIM28, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in wild-type and Dnmt3l KO ES cells. (F) DNMT3L and ZFP809-TRIM28 have a synergistic effect on the silencing of the proviral reporter and the methylation of the Mo-MuLV LTR DNAs. Wild-type (b1, b2, d1, and d2) and Dnmt3l KO (a1, a2, c1, and c2) 129S4/SvJae-derived ES cells were infected with either Mo-MuLVLUC (+PBS) (c1, c2, d1, and d2) or PBS-independent Mo-MuLVLUC/PBSQ (+PBSQ) (a1, a2, b1, and b2) viruses. We normalized the proviral luciferase activity to the relative virus copy numbers determined through quantitative PCR using primers specific to the PBS region of the exogenous Mo-MuLV as described previously (25) (Fig. 2D). The cells were cultured for the indicated periods, and luciferase activities were analyzed and normalized to the virus copy numbers revealed in panel F (left side). Data points are averages of three biological repeats; bars indicate SEMs. *, P < 0.05; **, P < 0.01. DNA methylation of the proviral LTRs 20 days postinfection is shown on the right side. DNA methylation percentage indicates the total number of black circles/total CpG sites. All P values were obtained by the nonparametric two-tailed Mann-Whitney test.