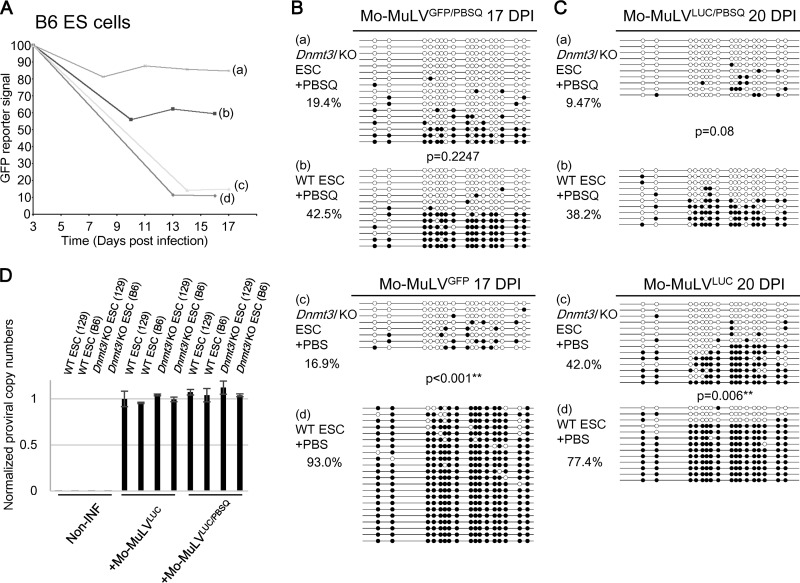
FIG 2.
DNMT3L- and ZFP809-TRIM28-mediated Mo-MuLV silencing in C57BL/6 background ES cells. (A) Wild-type and Dnmt3l KO ES cells in the C57BL/6 genetic background were sorted 3 days after Mo-MuLVGEP/PBS or Mo-MuLVGFP/PBSQ infection, and the GFP percentage was monitored. Wild-type (b and d) and Dnmt3l KO (a and c) C57BL/6-derived ES cells were infected with either Mo-MuLVGFP (c and d) or Mo-MuLVGFP/PBSQ (a and b) viruses for 3 days and then analyzed at the indicated time points by fluorescence-activated cell sorting (FACS) for GFP-positive ES cells. (B) DNA extracted and prepared for bisulfite analysis 17 days postinfection in the C57BL/6 genetic background. (C) Bisulfite sequencing of wild-type and Dnmt3l KO ES cells in the C57BL/6 genetic background after 20 days of Mo-MuLVLUC infection. DNA methylation percentage indicates the total number of black circles/total CpG sites. All P values were obtained by the nonparametric two-tailed Mann-Whitney test. **, P < 0.01. (D) qRT-PCR performed using primers specific to the PBS region of Mo-MuLV or internal control GAPDH after 2 days of Mo-MuLVLUC or Mo-MuLVLUC/PBSQ infection. Noninfected ES cells were used as a negative control. The values are averages of three biological repeats ± SEMs.