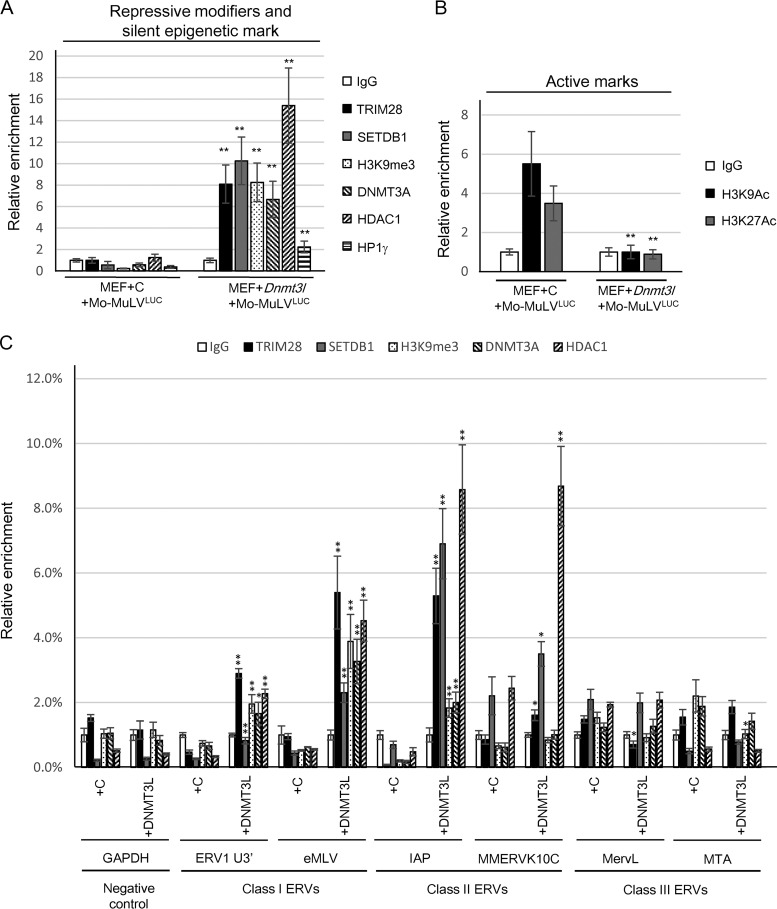
FIG 7.
DNMT3L can recruit epigenetic modifiers to induce repressive histone modifications on Mo-MuLV LTR and ERVs in MEFs. (A) DNMT3L induces TRIM28, SETDB1, DNMT3A, HDAC1, and HP1γ binding to proviral PBS sequence and increases H3K9me3 accumulation on the associated chromatin. Results of a chromatin immunoprecipitation (ChIP) assay of TRIM28, SETDB1, H3K9me3, DNMT3A, HDAC1, and HP1γ at the viral PBS sequence of control and DNMT3L-overexpressing MEFs after 2 days of Mo-MuLVLUC infection are shown. (B) DNMT3L decreases the active H3K9ac and H3K27ac marks on the proviral PBS sequence. (C) DNMT3L induces TRIM28, SETDB1, and HDAC1 binding to endogenous IAPs. Results of a ChIP assay of TRIM28, SETDB1, H3K9me3, DNMT3A, and HDAC1 at the ERVs are shown. ChIP using IgG antibody was used for background control. qRT-PCR was performed using sequence-specific primers ERV1 U3′ and endogenous MLV (class I), IAP and MMERVK10C (class II), and MervL and MTA (class III) as described previously (52). Primers specific for GAPDH served a negative control. Each ChIP experiment shows the mean enrichment ± the SEM from three biological repeats. Relative enrichment values (percentage of input) in all ChIP experiments were normalized to the total input of the samples. The relative enrichment values were compared statistically between control (+C) and DNMT3L-expressing (+DNMT3L) MEFs in each ERV primer set. Student's t tests were used for statistical analysis. *, P < 0.05; **, P < 0.01.