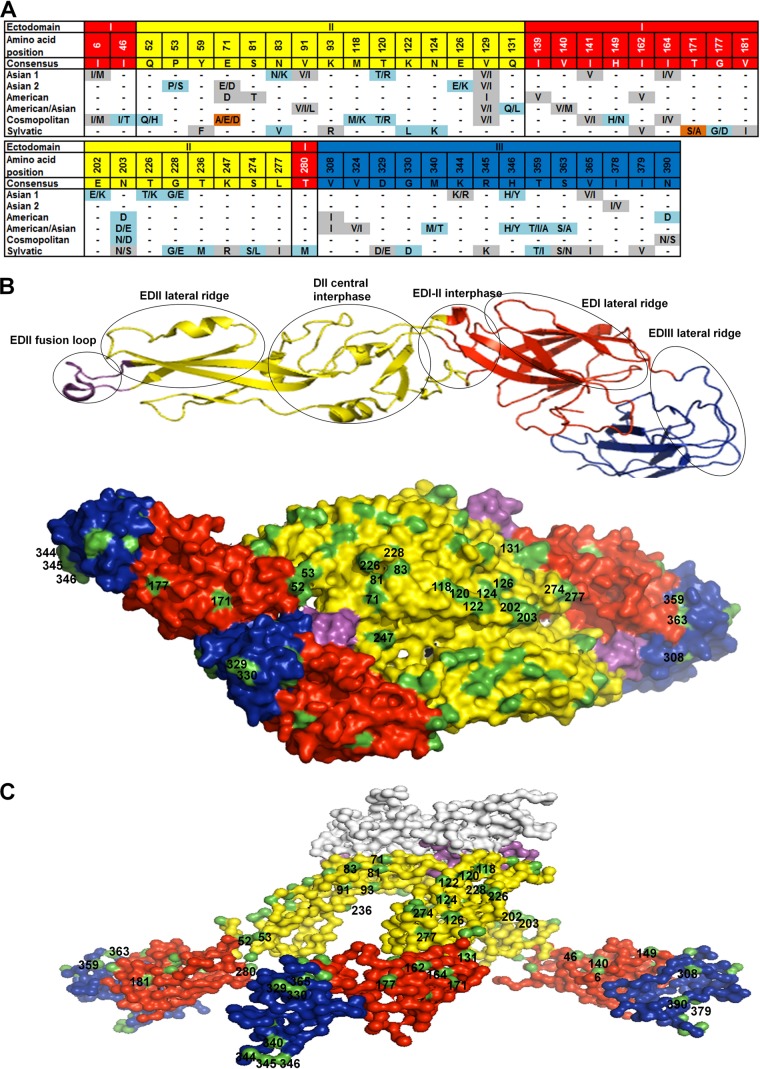
FIG 1.
Informative sites in the envelope protein of DENV-2 genotypes. (A) Alignment showing the variable amino acid positions and consensus sequence in the E-protein EDs. Gray shading, conservative amino acid substitutions; light blue shading, nonconservative substitutions; brown shading, nonconservative substitutions unique to certain genotypes. (B) Structural locations of surface-exposed informative sites. (Top) Ribbon diagram of the monomeric E protein with several of the known antigenic sites marked in circles; (bottom) top view of the dimeric E-protein crystal model of the mature DENV-2 particle (PDB accession number 3J27) depicted as a surface representation. (C) Side view surface representation of the model of the trimeric E-protein crystal structure of the immature DENV-2 particle (PDB accession number 3C6D). In panels A to C, EDI, EDII, EDIII, and the fusion loop are colored red, yellow, blue, and violet, respectively. Surface-exposed residues (B and C) are highlighted in green. The prM protein (C) is white.