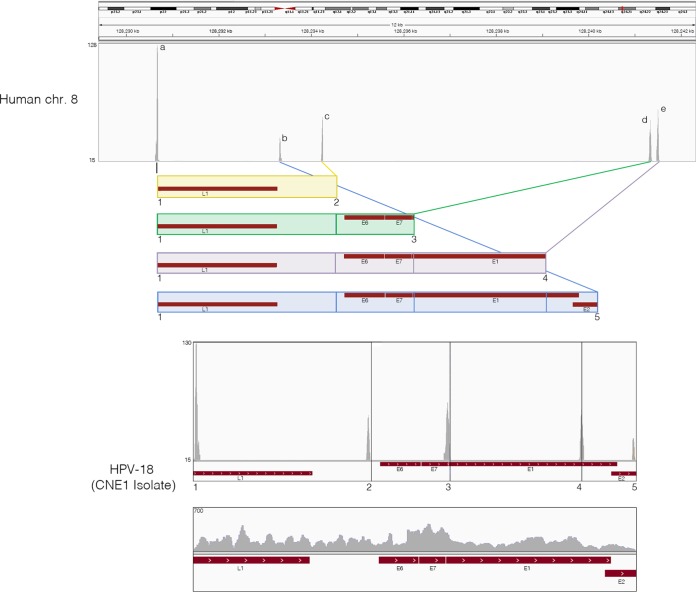
FIG 2.
HPV+ NPCs demonstrate same pattern of viral and cellular genomic rearrangement as HeLa cells. Strand-specific sequencing data from ribodepleted CNE1 RNA were analyzed for integration sites and breakpoints. First, 25-mer reads were extracted from both ends of each read and aligned to a reference genome containing both the human hg19 and HPV-CNE1 genomes. Next, fusion reads that were aligned to both human and HPV genomes were identified and visualized in the IGV genomic browser. Potential HPV integration patterns are represented as colored boxes. Read coverage peaks represent chromosome 8 integration sites (a to e) and HPV genome breakpoints (1 to 5). The rearranged HPV integrated genome is shown with breakpoints highlighted by numbers and corresponding read coverage.